Cell Ranger7.1, printed on 04/05/2025
In this tutorial, you will:
To follow along, you must:
The cellranger vdj pipeline takes FASTQ files from cellranger mkfastq,BCL Convert, or bcl2fastq for V(D)J libraries and performs sequence assembly and paired clonotype calling. It uses the Chromium cellular barcodes and UMIs to assemble V(D)J transcripts per cell. Clonotypes and CDR3 sequences are output as a .vloupe file which can be loaded into the Loupe V(D)J Browser.
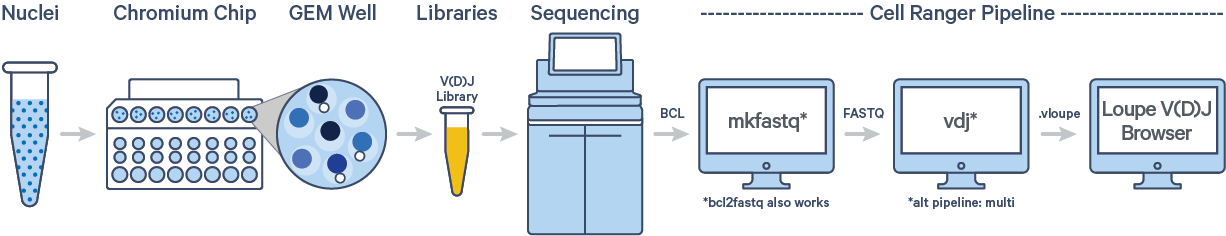
In this tutorial, we will work with the Human B cells dataset from a Healthy Donor (1k cells). This dataset was sequenced with the following configuration:
Paired-end (26X90), dual-indexed sequencing. Read 1: 26 cycles, i7 index: 10 cycles, i5 index: 10 cycles, Read 2: 90 cycles
Watch a short video tutorial or follow the text instructions below.
Open up a terminal window. You may log in to a remote server or choose to perform the compute on your local machine. Refer to the System Requirements page for details.
In the working directory, create a new folder called dataset-vdj-practice/ and cd into that folder:
mkdir dataset-vdj-practice/ cd dataset-vdj-practice/
Download the input FASTQ files:
curl -LO https://cf.10xgenomics.com/samples/cell-vdj/6.0.0/sc5p_v2_hs_B_1k_multi_5gex_b_Multiplex/sc5p_v2_hs_B_1k_multi_5gex_b_Multiplex_fastqs.tar
A file named sc5p_v2_hs_B_1k_multi_5gex_b_Multiplex_fastqs.tar should appear in your directory when you list files with the ls command.
Uncompress the FASTQs:
tar -xf sc5p_v2_hs_B_1k_multi_5gex_b_Multiplex_fastqs.tar
You should now see a folder called sc5p_v2_hs_B_1k_multi_5gex_b_fastqs that contains two subfolders sc5p_v2_hs_B_1k_5gex_fastqs and sc5p_v2_hs_B_1k_b_fastqs
Navigate back to the working directory:
cd ..
(Double check you are in the right place by running the ls command; the working directory should have a folder called sc5p_v2_hs_B_1k_multi_5gex_b_Multiplex_fastqs )
Download the V(D)J reference in the working directory:
curl -O https://cf.10xgenomics.com/supp/cell-vdj/refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0.tar.gz tar -xf refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0.tar.gz
Once you have all the necessary files, make a new directory called runs in your home directory:
mkdir runs/ cd runs/
You will run cellranger vdj in the runs/ directory.
After downloading the FASTQ files and a V(D)J reference transcriptome, you are ready to run cellranger vdj.
Print the usage statement to get a list of all the options:
cellranger vdj --help
The output should look similar to:
user_prompt$ cellranger vdj --help
cellranger-vdj
Assembles single-cell VDJ receptor sequences from 10x Immune Profiling libraries
USAGE:
cellranger vdj [FLAGS] [OPTIONS] --id
FLAGS:
--denovo Run in reference-free mode (do not use annotations)
--dry Do not execute the pipeline. Generate a pipeline
invocation (.mro) file and stop
--disable-ui Do not serve the web UI
--noexit Keep web UI running after pipestance completes or fails
--nopreflight Skip preflight checks
-h, --help Prints help information
OPTIONS:
--id
A unique run id and output folder name [a-zA-Z0-9_-]+
--description
Sample description to embed in output files [default: ]
--reference
Path of folder containing 10x-compatible VDJ reference. Optional if
'--denovo' is specified
--fastqs ... Path to input FASTQ data
--project
Name of the project folder within a mkfastq or bcl2fastq-generated
folder from which to pick FASTQs
....
| This tutorial assumes that the cellranger vdj run is being executed in the working-directory/runs/ directory. Maintaining the folder structure described in this tutorial is essential to execute code without path modifications |
| Option | Description |
|---|---|
| --id | The id argument must be a unique run ID. We will call this run HumanB_Cell based on the sample type in the example dataset. |
| --reference | a path that points at the V(D)J reference transcriptome. Path can be absolute or relative. In this example, the V(D)J reference is in the refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0/ directory located within the working directory. The relative path to the V(D)J reference from the runs directory should look like this ../refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0 |
| --fastqs | the fastqs argument requires a path to the directory containing FASTQ files. In this example, the FASTQs are in the dataset-vdj-practice/sc5p_v2_hs_B_1k_multi_5gex_b_fastqs/ directory. The relative path to this directory should be ../dataset-vdj-practice/sc5p_v2_hs_B_1k_multi_5gex_b_fastqs/sc5p_v2_hs_B_1k_b_fastqs |
| --sample | the sample name as specified in the sample sheet supplied to mkfastq. Typically, it is the prefix string common to all FASTQ files for one sample. In this example, the sample name should be sc5p_v2_hs_B_1k_b |
| --localcores | the localcores flag tells cellranger vdj to use a specified number of cores. We will use 8 cores in this example. |
| --localmem | the localmem flag specifies the amount of memory (in GB) for cellranger vdj to allocate to this run. We will ask for 64 GB for this example run. |
To learn more about all the cellranger vdj arguments, refer to the Running Pipelines section.
Watch a short video tutorial or follow the text instructions below.
Before running cellranger vdj, verify your directory structure:
. ├── dataset-vdj-practice │ └── sc5p_v2_hs_B_1k_multi_5gex_b_fastqs │ ├── sc5p_v2_hs_B_1k_5gex_fastqs │ └── sc5p_v2_hs_B_1k_b_fastqs ├── refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0 │ ├── fasta │ │ ├── regions.fa │ │ └── supp_regions.fa │ └── reference.json └── runs
From within the working-directory/runs/ directory, run cellranger vdj. If you have the same directory structure as this tutorial, you should be able to copy and paste this code into your command prompt (no need to type it out!). If you need to change file paths, the easiest way to customize code is by first pasting it into a text editor (e.g. TextEdit for MacOS). Then, you can copy and paste the customized code into your command prompt.
cellranger vdj --id=HumanB_Cell \
--reference=../refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0 \
--fastqs=../dataset-vdj-practice/sc5p_v2_hs_B_1k_multi_5gex_b_fastqs/sc5p_v2_hs_B_1k_b_fastqs \
--sample=sc5p_v2_hs_B_1k_b \
--localcores=8 \
--localmem=64 \
Refer to the Set up section above to understand the specific flags.
The run begins similar to this:
Serving UI at http://bespin1.fuzzplex.com:45033?auth=o5bjTf5rSJ1HDx-hX-bIBcMS4_7w9agaXXFcDNj1ulY Running preflight checks (please wait)... Checking sample info... Checking FASTQ folder... Checking reference... Checking reference_path (working-directory/refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0) Checking inner enrichment primers... Checking chain... mrc: v4.0.6 mrp: v4.0.6 Anaconda: Python 3.8.2 numpy: 1.19.2 scipy: 1.6.2 pysam: 0.16.0.1 h5py: 3.2.1
When the output of the cellranger vdj command says, “Pipestance completed successfully!”, the job is done:
Outputs:
- Run summary HTML: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/web_summary.html
- Run summary CSV: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/metrics_summary.csv
- Clonotype info: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/clonotypes.csv
- Filtered contig sequences FASTA: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/filtered_contig.fasta
- Filtered contig sequences FASTQ: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/filtered_contig.fastq
- Filtered contigs (CSV): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/filtered_contig_annotations.csv
- All-contig FASTA: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig.fasta
- All-contig FASTA index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig.fasta.fai
- All-contig FASTQ: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig.fastq
- Read-contig alignments: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig.bam
- Read-contig alignment index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig.bam.bai
- All contig annotations (JSON): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig_annotations.json
- All contig annotations (BED): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig_annotations.bed
- All contig annotations (CSV): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/all_contig_annotations.csv
- Barcodes that are declared to be targetted cells: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/cell_barcodes.json
- Clonotype consensus FASTA: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/consensus.fasta
- Clonotype consensus FASTA index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/consensus.fasta.fai
- Contig-consensus alignments: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/consensus.bam
- Contig-consensus alignment index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/consensus.bam.bai
- Clonotype consensus annotations (CSV): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/consensus_annotations.csv
- Concatenated reference sequences: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/concat_ref.fasta
- Concatenated reference index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/concat_ref.fasta.fai
- Contig-reference alignments: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/concat_ref.bam
- Contig-reference alignment index: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/concat_ref.bam.bai
- Loupe V(D)J Browser file: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/vloupe.vloupe
- V(D)J reference:
fasta:
regions: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/vdj_reference/fasta/regions.fa
donor_regions: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/vdj_reference/fasta/donor_regions.fa
reference: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/vdj_reference/reference.json
- AIRR Rearrangement TSV: /mnt/deck7/working-directory/runs/HumanB_Cell/outs/airr_rearrangement.tsv
- All contig info (ProtoBuf format): /mnt/deck7/working-directory/runs/HumanB_Cell/outs/vdj_contig_info.pb
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
Watch a short video tutorial or follow the text instructions below.
A successful cellranger vdj run produces a new directory called HumanB_Cell/ (based on the --id flag specified during the run). The contents of the HumanB_Cell/ directory:
── runs
└── HumanB_Cell
├── _cmdline
├── _filelist
├── _finalstate
├── _invocation
├── _jobmode
├── _log
├── _mrosource
├── outs
├── _perf
├── SC_VDJ_ASSEMBLER_CS
├── _sitecheck
├── _tags
├── _timestamp
├── _uuid
├── vdj_test.mri.tgz
├── _vdrkill
└── _versions
The outs/ directory contains all important output files generated by the cellranger vdj pipeline.
── runs
└── HumanB_Cell
└──outs
├── airr_rearrangement.tsv
├── all_contig_annotations.bed
├── all_contig_annotations.csv
├── all_contig_annotations.json
├── all_contig.bam
├── all_contig.bam.bai
├── all_contig.fasta
├── all_contig.fasta.fai
├── all_contig.fastq
├── cell_barcodes.json
├── clonotypes.csv
├── concat_ref.bam
├── concat_ref.bam.bai
├── concat_ref.fasta
├── concat_ref.fasta.fai
├── consensus_annotations.csv
├── consensus.bam
├── consensus.bam.bai
├── consensus.fasta
├── consensus.fasta.fai
├── filtered_contig_annotations.csv
├── filtered_contig.fasta
├── filtered_contig.fastq
├── metrics_summary.csv
├── vdj_contig_info.pb
├── vdj_reference
│ ├── fasta
│ │ ├── donor_regions.fa
│ │ └── regions.fa
│ └── reference.json
├── vloupe.vloupe
└── web_summary.html