Cell Ranger, printed on 03/30/2025
Loupe V(D)J Browser 4.0 has been updated to support changes to Cell Ranger 5.0, including
V(D)J aggr and the new clonotype grouping algorithm.
Loupe V(D)J Browser 4.0 is a required upgrade for processing data generated by the Cell
Ranger 5.0 pipeline. Due to extensive changes to the underlying infrastructure, .vloupe
files generated by Cell Ranger 4.0 and earlier Cell Ranger versions cannot be read into
Loupe V(D)J Browser 4.0 or Loupe Browser 5.0.
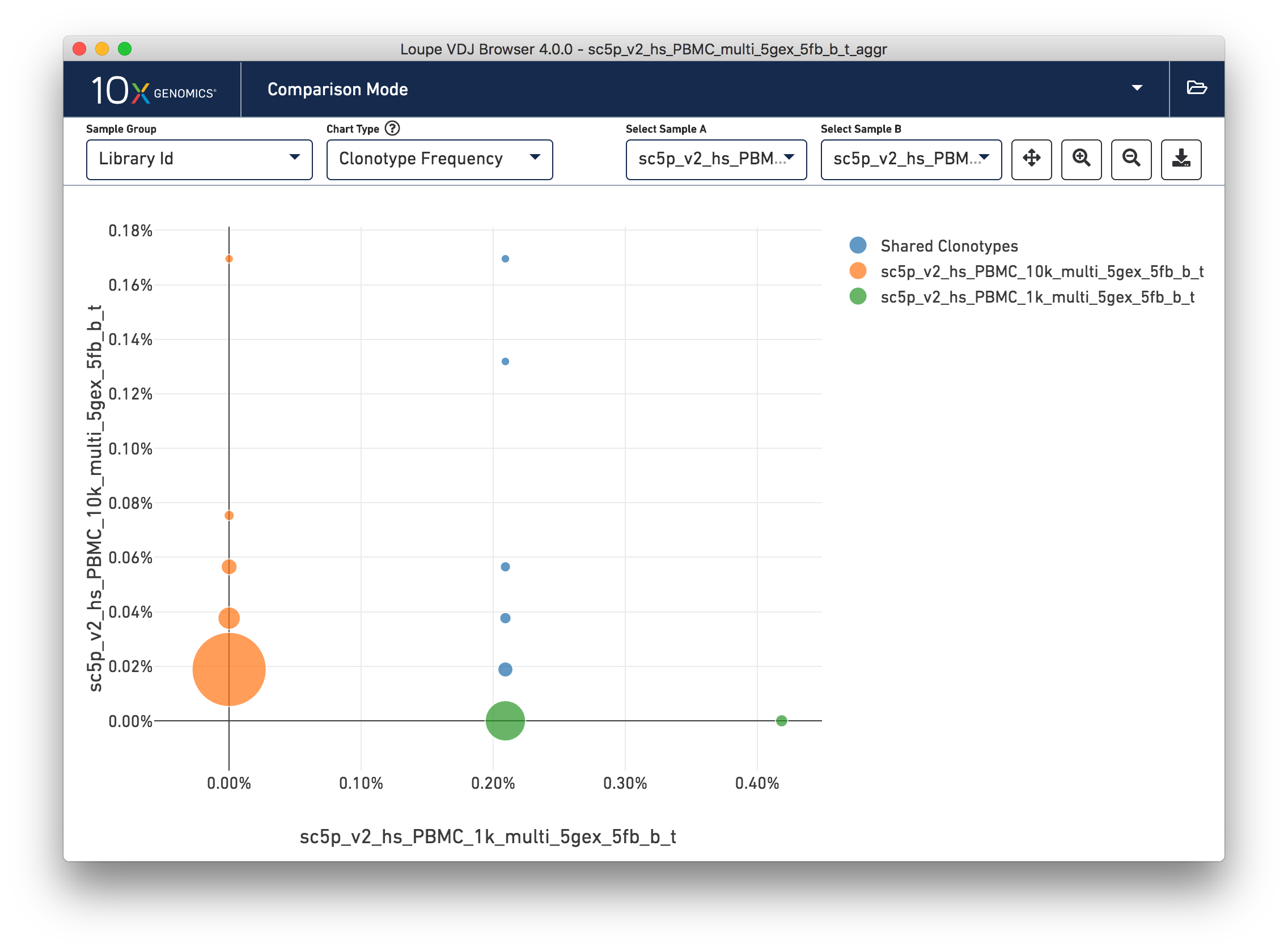
Loupe V(D)J Browser 4.0 now allows for comparison of samples in files generated by the
V(D)J aggr pipeline. Data can be divided into samples based on the columns in the
aggregation CSV provided to the aggr pipeline, such as donor, origin, library ID, and
any custom columns. A sample group dropdown has been added to all comparison mode charts,
allowing for quick navigation.
For .vloupe files with a large number of samples, a new selection option will appear to
enable displaying a subset of samples in a single view.
The clonotype frequency view is no longer accessible via a button in the sample table, but can still be found in the chart type dropdown. Selectors have now been added to the clonotype frequency chart to allow for quick switching between samples.
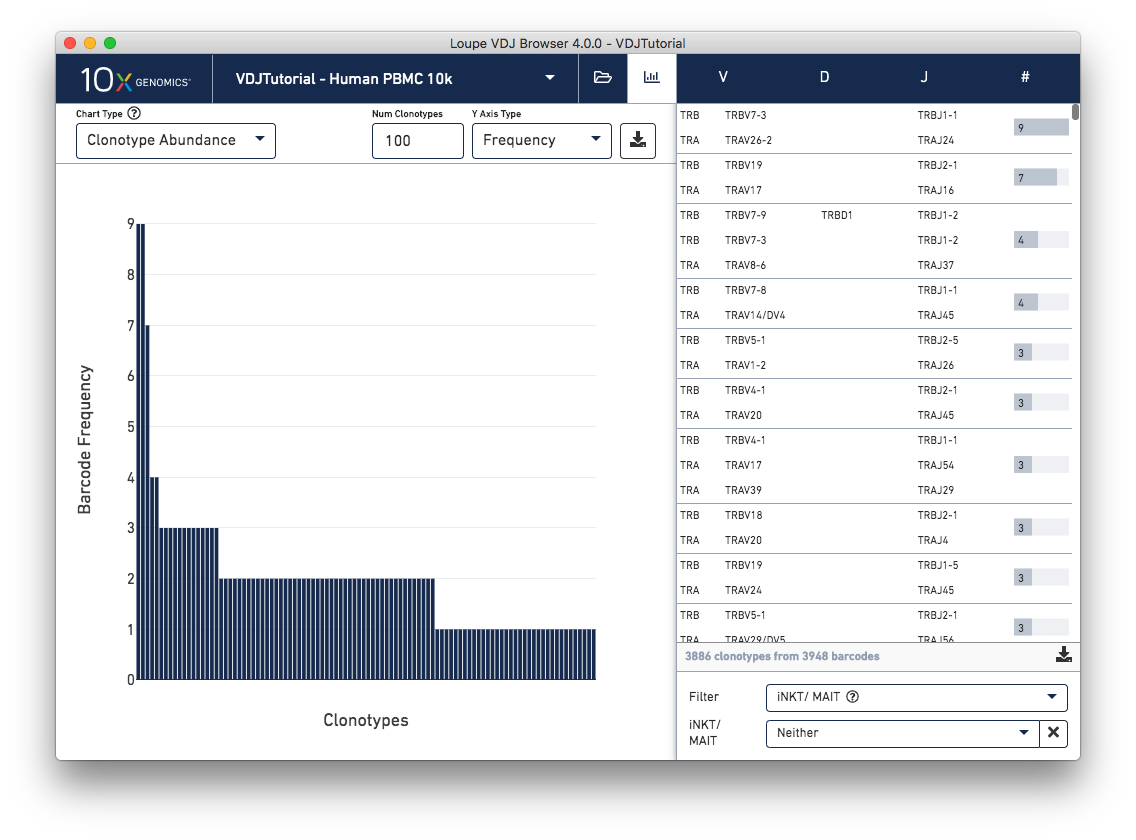
Identify potential invariant natural killer T (iNKT) cells and mucosal-associated invariant T (MAIT) cells with a new clonotype table filter. The iNKT/MAIT evidence filter can be used to view clonotypes that contain any cells with supporting evidence in any of the TRA genes, TRA junctions, TRB genes, or TRB junctions of being an iNKT or MAIT cell. This filter is only visible for T cell datasets.
There are a number of other powerful filtering options added in this release:
.vloupe files generated by
aggr.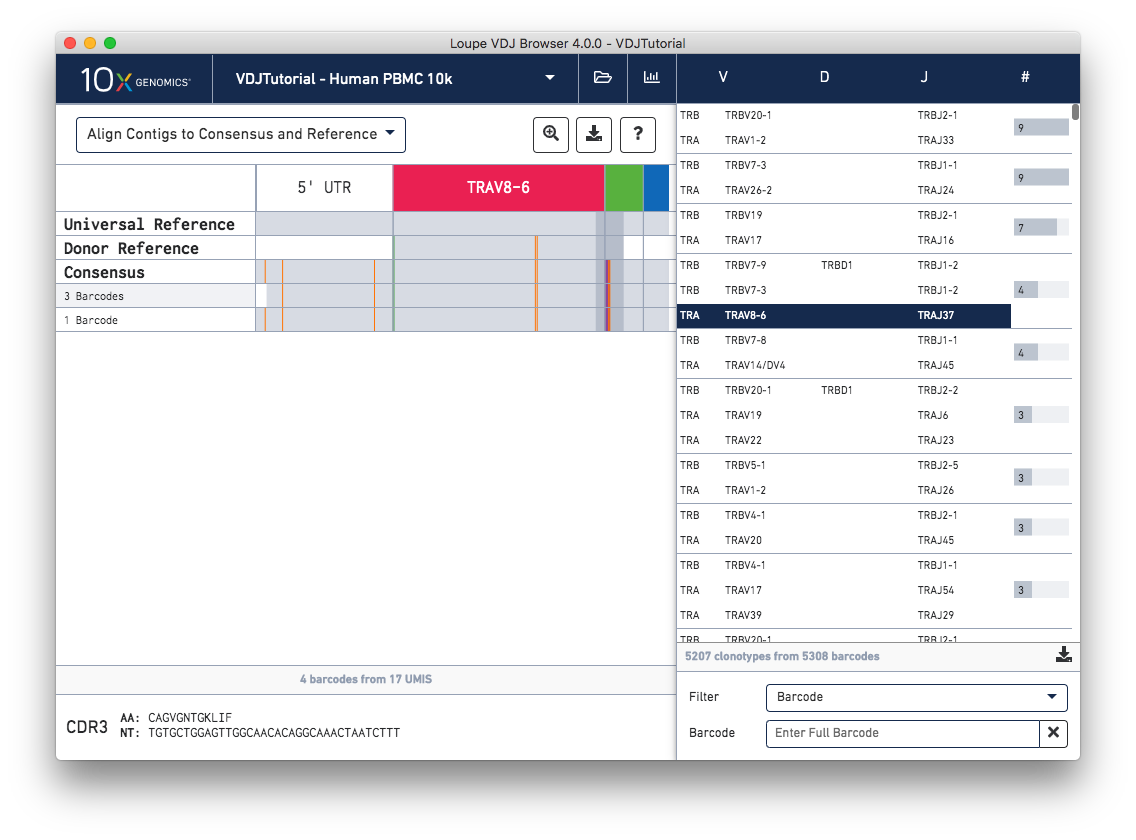
It’s now easier than ever to gain insights from the chain view. Rows in the chain view now represent exact subclonotypes instead of individual cells, meaning cells with identical immune receptor nucleotide sequences are now collapsed into a single row in the table. A Download Barcode List option is available when hovering over an exact subclonotype to see the full list of barcodes included in the row. Additionally, a donor reference is now visible between the universal reference and consensus. The donor reference only spans the length of the V gene.