Cell Ranger7.1, printed on 11/26/2024
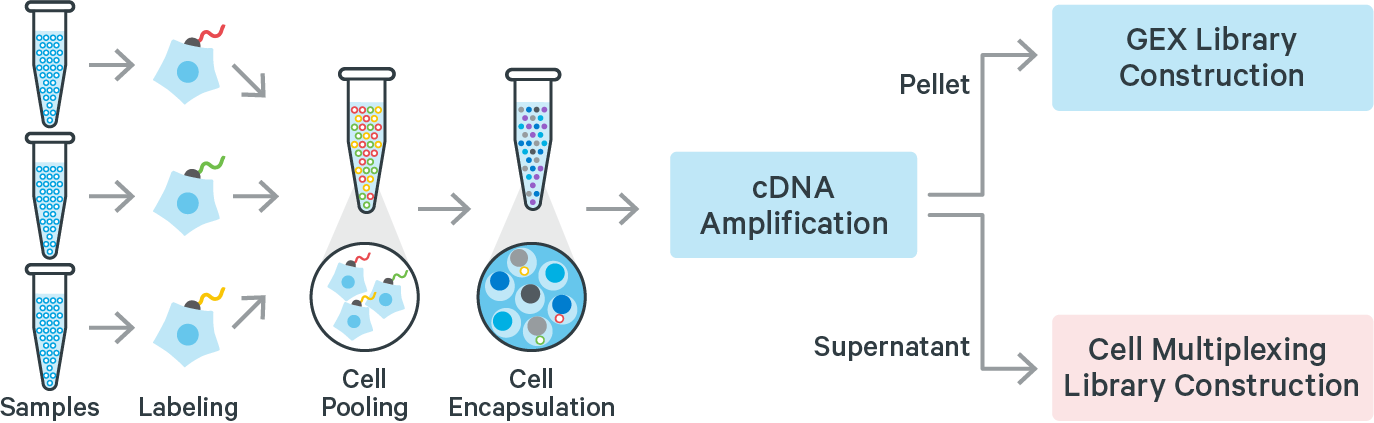
Cell Multiplexing refers to the labeling of a cell or nuclei sample with a molecular tag and subsequently mixing this sample with other labeled samples. This set of multiplexed samples can be processed together in a single GEM well. After cell encapsulation, library preparation, and sequencing, molecular tag information can be assigned to cells. Tag assignment enables identification of droplets that originally contained one (singlet) or more cells (multiplets). Cells assigned a given single tag are binned together, bioinformatically recapitulating the individual samples originally mixed together.
Key advantages of Cell Multiplexing include:
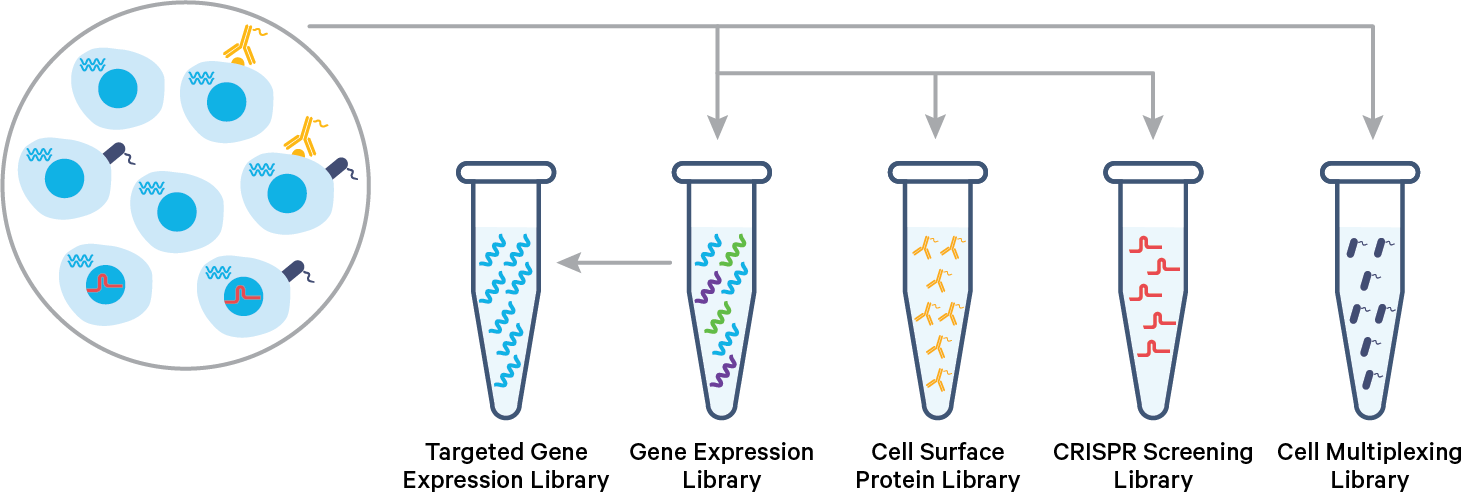
The 10x Genomics 3’ CellPlex Multiplexing Solution is a Feature Barcode technology, similar to existing 10x Genomics Cell Surface Protein and CRISPR Screening assays. 3’ CellPlex is enabled by SingleCell 3’ v3.1 gelbeads and reagents. Cells or nuclei labeled with Cell Multiplexing Oligos (CMOs) are pooled prior to loading onto a 10x Genomics chip. cDNA from poly-adenylated mRNA and DNA from the CMO Feature Barcode are generated simultaneously from the same single cells inside of a Gel Bead-in-Emulsion (GEM). 3’ CellPlex reagents are compatible with samples whose cell surface proteins have been labeled with TotalSeq-B antibody-oligonucleotide conjugates or samples transduced with Feature Barcode technology compatible sgRNA constructs.
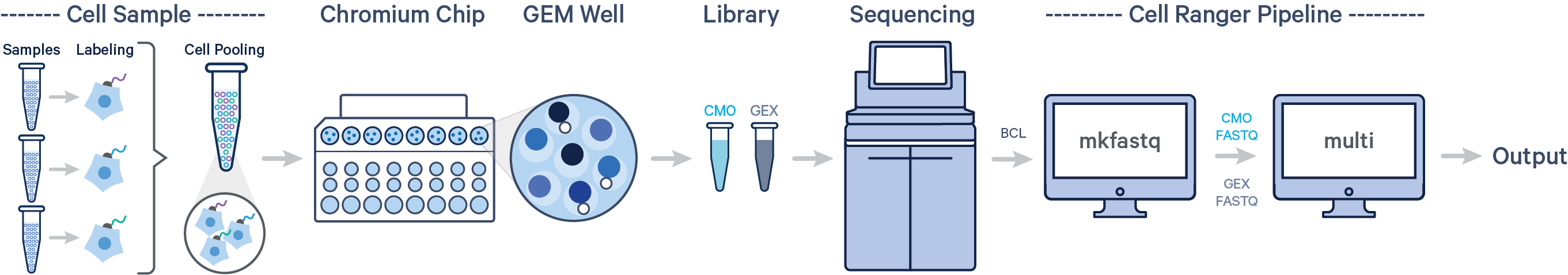
Cell Ranger v6.0 or later is required to analyze Cell Multiplexing data. Multiple samples are uniquely tagged with CMOs prior to pooling in a single GEM well, resulting in a CMO and Gene Expression (GEX) library for each GEM well. After demultiplexing the BCL files with cellranger mkfastq, run the cellranger multi pipeline on the combined FASTQ data for the CMO and GEX libraries to obtain separate per-sample output files for each CMO.