Cell Ranger7.1, printed on 03/29/2025
Cell Ranger v7.0 and later supports analyzing Fixed RNA Profiling (FRP) data with the cellranger multi pipeline. Cell Ranger v7.1 supports analysis of single cell FFPE (scFFPE) datasets.
First, follow the instructions on running cellranger mkfastq to generate FASTQ files. For example, if the flow cell ID was HAWT7ADXX, then cellranger mkfastq will output FASTQ files in HAWT7ADXX/outs/fastq_path. If you are already starting with FASTQ files, you can skip this step and proceed directly to run cellranger multi.
Running cellranger multi requires a config CSV, described below, invoking the following arguments:
| Argument | Description |
|---|---|
--id | A unique run ID string: e.g. sample345 that is also the output folder name. Cannot be more than 64 characters. |
--csv | Path to config CSV file with input libraries and analysis parameters. |
The multi config CSV contains both the library definitions and experimental design variables. The required sections differ slightly for analysis with single-sample ("singleplex") vs. multiplexed configurations. It is composed of up to four sections for FRP data:
Example formats for different product configurations are below. A template for a multi config CSV can be downloaded here or via the command line. Example multi config CSVs can be downloaded from 7.0 public datasets here.
| Multi Config CSV | |
|---|---|
Section: [gene-expression] | |
| Field | Description |
reference
| Required. Path of folder containing 10x Genomics-compatible genome reference. |
probe-set
| Required. Path to the probe set reference CSV file. This file is included with the Cell Ranger v7.0+ package and on the Download Links page. |
filter-probes
| Optional. Include all non-deprecated probes listed in the probe set reference CSV file. Probes that are predicted to have off-target activity to homologous genes are excluded from analysis by default. Setting filter-probes to false will result in UMI counts from all non-deprecated probes, including those with predicted off-target activity, to be used in the analysis. Probes whose ID is prefixed with DEPRECATED are always excluded from the analysis. Default: true
|
r1-length
| Optional. Limit the length of the input Read 1 sequence of Gene Expression libraries to the first N bases, where N is a user-supplied value. Note that the length includes the 10x Barcode and UMI sequences so do not set this below 26. This and r2-length are useful options for determining the optimal read length for sequencing. Default: do not trim Read 1. |
r2-length
| Optional. Limit the length of the input Read 2 sequence of Gene Expression libraries to the first N bases, where N is a user-supplied value. Trimming occurs before sequencing metrics are computed and therefore, limiting the length of Read 2 may affect Q30 scores. Default: do not trim Read 2. |
chemistry
| Optional. Assay configuration. NOTE: by default, the assay configuration is detected automatically (recommended mode). Users will typically not need to specify a chemistry. Options are: auto for auto-detection, or to override: SFRP for singleplex FRP and MFRP for multiplex FRP. Default: auto |
expect-cells
| Optional. Override the pipeline’s auto-estimate. See cell calling algorithm overview for details on how this parameter is used. If used, enter the expected number of recovered cells. Specifying library-level expect-cells in the [gene-expression] section is only valid for the singleplex FRP configuration; note this flag name has a dash (-).
|
force-cells
| Optional. Force pipeline to use this number of cells, bypassing cell detection. Specifying library-level force-cells in the [gene-expression] section is only valid for the singleplex Fixed RNA Profiling configuration; note this flag name has a dash (-). Default: detect cells using Cell Ranger's cell calling algorithm |
no-secondary
| Optional. Disable secondary analysis, e.g. clustering. Default: false |
no-bam
| Optional. Set this flag to true to skip BAM file generation. If unsure, we recommend setting this option to true for Fixed RNA Profiling analysis, which will reduce the total computation time for the pipestance and the size of the output directory. Default: false |
check-library-compatibility
| Optional.This option allows users to disable the check that evaluates 10x Barcode overlap between libraries when multiple libraries are specified (e.g., Gene Expression + Antibody Capture). Setting this option to false will disable the check across all library combinations. We recommend running this check (default), however if the pipeline errors out, users can bypass the check to generate outputs for troubleshooting. Default: true |
Section: [feature] | |
| Field | Description |
reference
| Path to the Feature reference CSV file, declaring Antibody Capture constructs and associated barcodes. Required for Feature Barcode libraries, otherwise optional. |
r1-length
| Optional. Limit the length of the input Read 1 sequence of Feature Barcode libraries to the first N bases, where N is a user-supplied value. Note that the length includes the 10x Barcode and UMI sequences so do not set this below 26. This and r2-length are useful options for determining the optimal read length for sequencing. Default: do not trim Read 1. |
r2-length
| Optional. Limit the length of the input Read 2 sequence of Feature Barcode libraries to the first N bases, where N is a user-supplied value. Trimming occurs before sequencing metrics are computed and therefore, limiting the length of Read 2 may affect Q30 scores. Default: do not trim Read 2. |
Section: [libraries] (see also Specifying Input FASTQ Files for cellranger multi) | |
| Column | Description |
fastq_id
| Required. The Illumina sample name to analyze. This will be as specified in the sample sheet supplied to mkfastq or bcl2fastq. |
fastqs
| Required. Path to the folder containing the FASTQ files to be analyzed. Generally, this will be the fastq_path folder generated by cellranger mkfastq. |
feature_types
| Required. The underlying feature type of the library, which must be one of Gene Expression or Antibody Capture. |
lanes
| Optional. The lanes associated with this sample, separated with a pipe (e.g., 1|2). Defaults to using all lanes. |
physical_library_id
| Optional. Library type. NOTE: by default, the library type is detected automatically based on specified feature_types (recommended mode). Users typically do not need to include the physical_library_id column in the CSV file. |
subsample_rate
| Optional. The rate at which reads from the provided FASTQ files are sampled. Must be strictly greater than 0 and less than or equal to 1. |
Section: [samples] | |
| Column | Description |
sample_id
| Required. A name to identify a multiplexed sample or a single sample with multiple Probe Barcodes. Must be alphanumeric with hyphens and/or underscores, and less than 64 characters. |
probe_barcode_ids
| Required. The Fixed RNA Probe Barcode IDs used for this sample. If multiple Probe Barcodes were used for a sample, separate IDs with a pipe (e.g., BC001|BC002). |
description
| Optional. A description for the sample. |
expect_cells
| Optional. Override the pipeline’s auto-estimate. See Gene Expression algorithm overview for details. If used, enter the expected number of recovered cells. Specifying sample-level expect_cells in the [samples] section is only valid for the multiplex Fixed RNA Profiling configuration; note this column name has an underscore (_).
|
force_cells
| Optional. Force pipeline to use this number of cells, bypassing cell detection. Specifying sample-level force_cells in the [samples] section is only valid for the multiplex Fixed RNA Profiling configuration; note this column name has an underscore (_). Default: detect cells using EmptyDrops |
cd /home/jdoe/runs cellranger multi --id=sample345 --csv=/home/jdoe/sample345.csv
Martian Runtime - v4.0.8 Running preflight checks (please wait)... ...
By default, Cell Ranger will use all of the cores available on your
system to execute pipeline stages. You can specify a different number of cores
to use with the --localcores option; for example, --localcores=16
will limit Cell Ranger to using up to sixteen cores at once. Similarly,
--localmem will restrict the amount of memory (in GB) used by
Cell Ranger.
The pipeline will create a new folder named with the run ID you specified using the --id argument (e.g. /home/jdoe/runs/sample345) for its output. If this folder already exists, Cell Ranger will assume it is an existing pipestance and attempt to resume running it.
Waiting 6 seconds for UI to do final refresh. Pipestance completed successfully! yyyy-mm-dd hh:mm:ss Shutting down. Saving pipestance info to "tiny/tiny.mri.tgz"
sample345). The subfolder named outs/ will contain the main pipeline outputs.Cell Ranger v7.1 enables users to download a multi config CSV template by running:
cellranger multi-template --output=/path/to/FILE.csv
Remember to replace code in red with the path to the directory in which you wish to output the template. Omitting the file path downloads the file into your working directory. After downloading, please remember to customize the template based on your assay and experimental design.
To print a list and description of all configurable parameters available in cellranger multi, run
cellranger multi-template --parameters
Specifying both --parameters and --output will output a parameter documentation file. Run cellranger multi-template --help or cellranger multi-template -h for more information about available flags.
Here are a few example multi config CSVs for some common Fixed RNA Profiling (FRP) assay configurations, along with simplified diagrams for the corresponding experimental set up. Make sure to replace /path/to with the actual full path to your data, and edit any text in red according to the experiment's sample/library/file names.
Important: In the examples below, we set no-bam to “true” so Cell Ranger will not generate a BAM file. This setting is recommended for Fixed RNA Profiling libraries and will reduce both the total computation time for the pipestance and the size of the output directory.
| Experimental Design | Multi config CSV |
Singleplex FRP, 1 Probe Barcode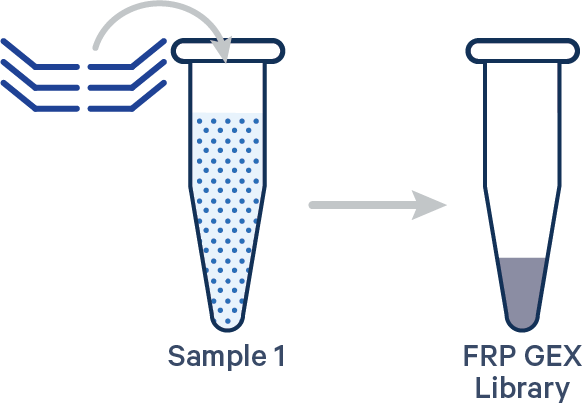 See example dataset Note: this library configuration does not use the [samples] section in the multi config CSV.
|
[gene-expression] reference,/path/to/transcriptome probe-set,/path/to/probe/set no-bam,true #do not generate BAM file |
Singleplex FRP with Antibody Capture, 1 Probe Barcode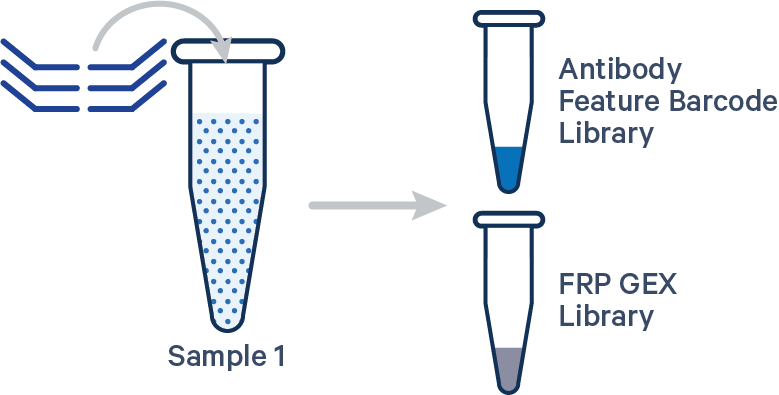 See example dataset Antibody Capture is compatible in this configuration. There is one sample, one Probe Barcode, and two libraries (Gene Expression and Antibody Capture). Note: this library configuration does not use the [samples]section in the multi config CSV. |
[gene-expression] reference,/path/to/transcriptome probe-set,/path/to/probe/set no-bam,true #do not generate BAM file |
|
Multiplex FRP, multiple Probe Barcodes/sample A. Multiple biological samples 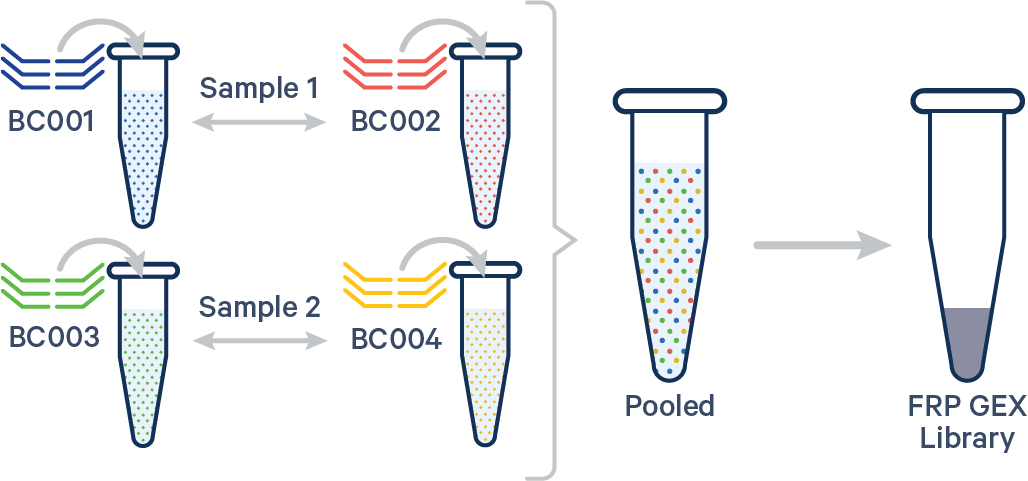 |
[gene-expression] reference,/path/to/transcriptome probe-set,/path/to/probe/set no-bam,true #do not generate BAM file |
|
B. Single biological sample In this case, the config CSV must include a [samples] section to specify the Probe Barcodes since two Probe Barcodes were used for a single sample in this experiment. Note that this configuration is not compatible with Antibody Capture. |
[gene-expression] reference,/path/to/transcriptome probe-set,/path/to/probe/set no-bam,true #do not generate BAM file |
Multiplex FRP, 1 Probe Barcode/sample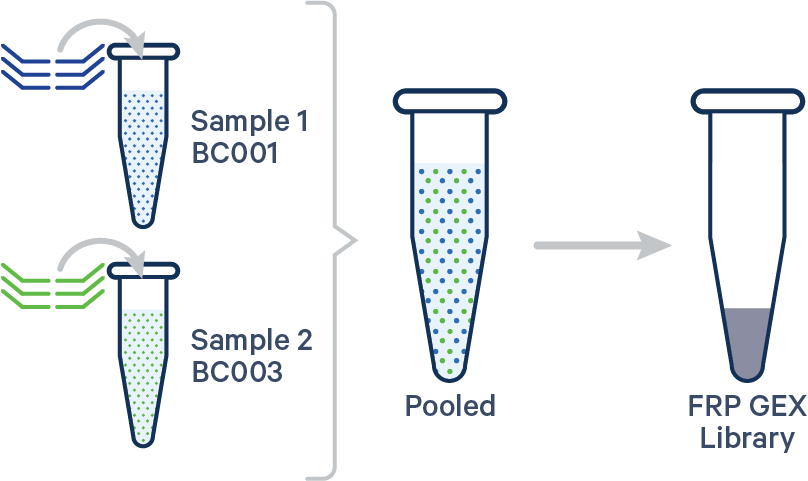 See example dataset Note that this configuration is not compatible with Antibody Capture. |
[gene-expression] reference,/path/to/transcriptome probe-set,/path/to/probe/set no-bam,true #do not generate BAM file |
The v1.0.1 probe set reference CSV files for human and mouse can be found in the probe_sets directory of the Cell Ranger package (v7.1):
cellranger-7.1.0/probe_sets/ └── Chromium_Human_Transcriptome_Probe_Set_v1.0.1_GRCh38-2020-A.csv └── Chromium_Mouse_Transcriptome_Probe_Set_v1.0.1_mm10-2020-A.csv
region column). |
The v1.0.1 and v1.0 probe set reference CSV files and additional support files for human and mouse:
The following Gene Expression (GEX) assay chemistry combinations may be aggregated with cellranger aggr, and are either supported (validated by 10x Genomics) or possible (will run, but not the validated intended usage). Targeted Gene Expression is not compatible for aggregation with Fixed RNA Gene Expression (FRP). Some assay chemistry combinations are supported for aggregation as long as they have the same feature reference CSV file (+) and/or probe set reference CSV file (^); please see this FAQ page for details about aggregating these assay combinations.
| Assay chemistry | Singleplex FRP | Singleplex FRP, Antibody | Multiplex FRP |
|---|---|---|---|
| Singleplex FRP | Supported^ | - | - |
| Singleplex FRP, Antibody | Supported+^ | Supported+^ | - |
| Multiplex FRP | Supported^ | Supported+^ | Supported^ |
| 3' or 5' GEX library | Possible | Possible+^ | Possible |