Space Ranger2.0, printed on 03/25/2025
| The CytAssist faciliated workflow now supports Fresh Frozen (FF) and Fixed Frozen (FxF) samples using probe-based whole transcriptome profiling. |
Space Ranger's pipelines analyze sequencing data produced from Visium Spatial Gene Expression for Formalin Fixed Paraffin Embedded (FFPE) and Fresh Frozen (FF) tissue samples. The CytAssist instrument aids in the transfer of analytes from the FFPE or FF tissue section on a standard glass slides to the CytAssist Spatial Gene Expression Slide. The workflow diagram shows all the required inputs and the corresponding spaceranger flags for analyzing CytAssist enabled FFPE, FF or Fixed Frozen (FxF) tissue samples:
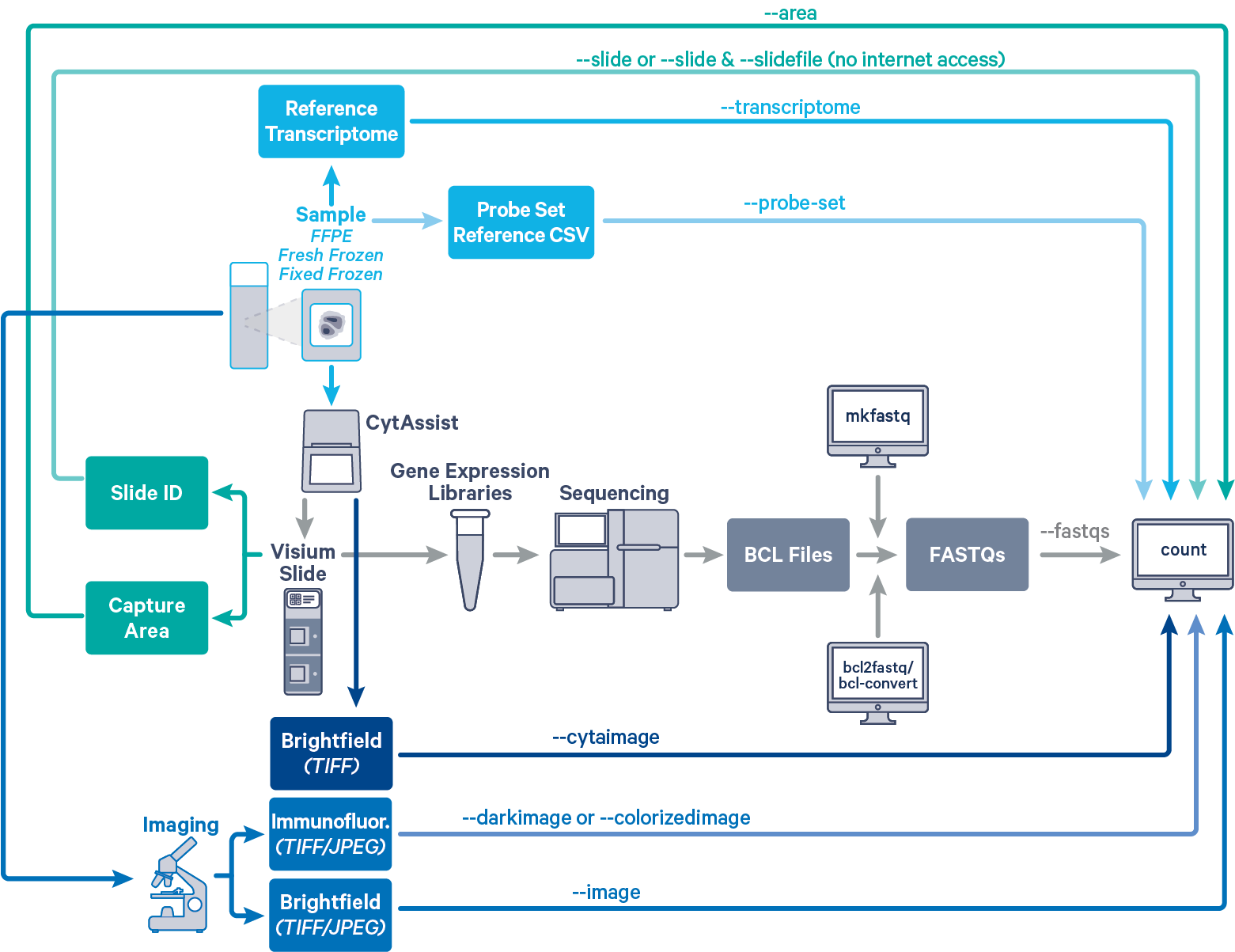
The spaceranger count is run on each individual Capture Area in the Visium slide. The required inputs are
--fastqs)
TIFF with optional microscope image in either TIFF, QPTIFFor JPEG format
--cytaimage for image captured by CytAssist instrument (contains fiducial frame)--image for brightfield microscope image--darkimage for dark background fluorescence microscope image--colorizedimage for composite colored fluorescence microscope image--slide & --area if spaceranger has access to internet--slidefile, --slide & --area if spaceranger has no access to internet. The slide layout file is directly downloaded--unknown-slide if Visium slide details are unknown--transcriptome)--probe-set flagFor a list of accepted arguments, see the Command Line Argument Reference below, or run spaceranger count --help.
Select the tab corresponding to the imaging workflow.
There are two possible workflow variations depending on whether or not a high resolution microscopy image of the tissue section on the standard glass is supplied. Note that success of the automatic image processing pipeline is based on the assumption that the tissue morphology and overall shape on the microscope image matches that captured by CytAssist. The presence of tissue folds, tears, and other artifacts that prevent the CytAssist image from representing the tissue on the slide can lead to potential problems with pipeline processing.
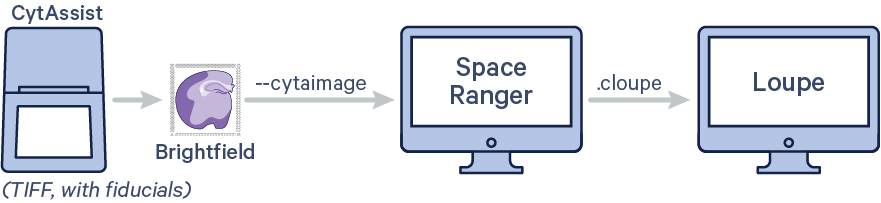
To generate spatial feature counts for a single library using automatic fiducial alignment and tissue detection on CytAssist image input, run spaceranger count with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --probe-set=/home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv\ #Path to Probe Set --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --cytaimage=/home/jdoe/runs/images/A1_sample345.tiff \ #Path to CytAssist image input --slide=V49J01-123 \ #Slide ID --area=A1 \ #Capture area --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmode
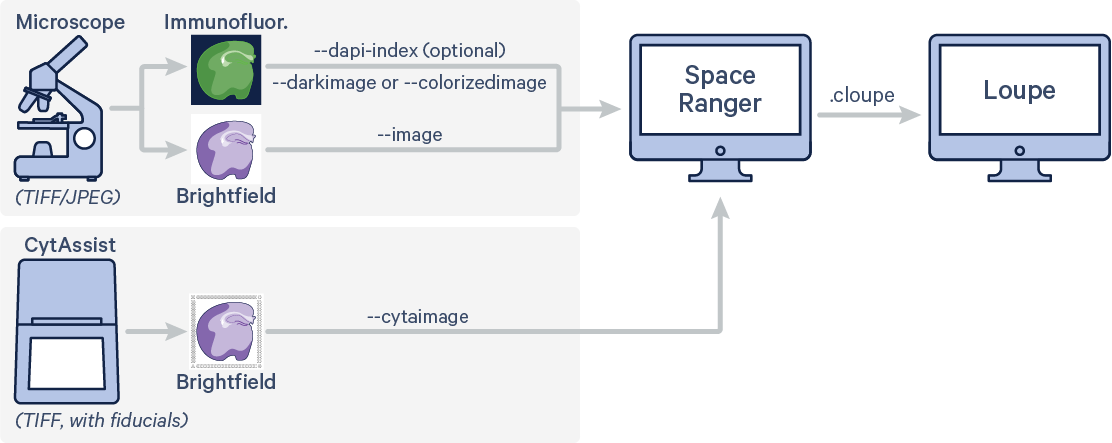
The example code here represents use of a multi channel immunofluorescence microscope image (IF) with one channel stained with DAPI in addition to CytAssist image. Run spaceranger count with the following arguments. Space Ranger will perform automatic image registration of the two image inputs as well as fiducial alignment and tissue detection on CytAssist image. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --probe-set=/home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv\ #Path to Probe Set --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --cytaimage=/home/jdoe/runs/images/A1_sample345.tiff \ #Path to CytAssist image input --darkimage=/home/jdoe/runs/images/IF_sample345.tiff \ #Path to IF microscope image input --dapi-index=2 \ #Numeric value corresponding to the DAPI channel --slide=V49J01-123 \ #Slide ID --area=A1 \ #Capture area --localcores=8 \ #Allowed cores in local mode --localmem=64 #Allowed memory (GB) in local mode
There are three possible workflow variations, depending on whether or not a high resolution microscopy image of the tissue section on the standard glass slide was supplied, and the types of manual alignment performed in Loupe Browser. The following examples use Brightfield microscope image. The arguments to specify the fluorescence image can be either --darkimage or --colorizedimage depending on the image format. Note that the brightfield CytAssist image, which has the fiducial frame, is used for fiducial alignment and tissue detection workflow. Details about the different supported fluorescence image formats are described in Image Recommendations.
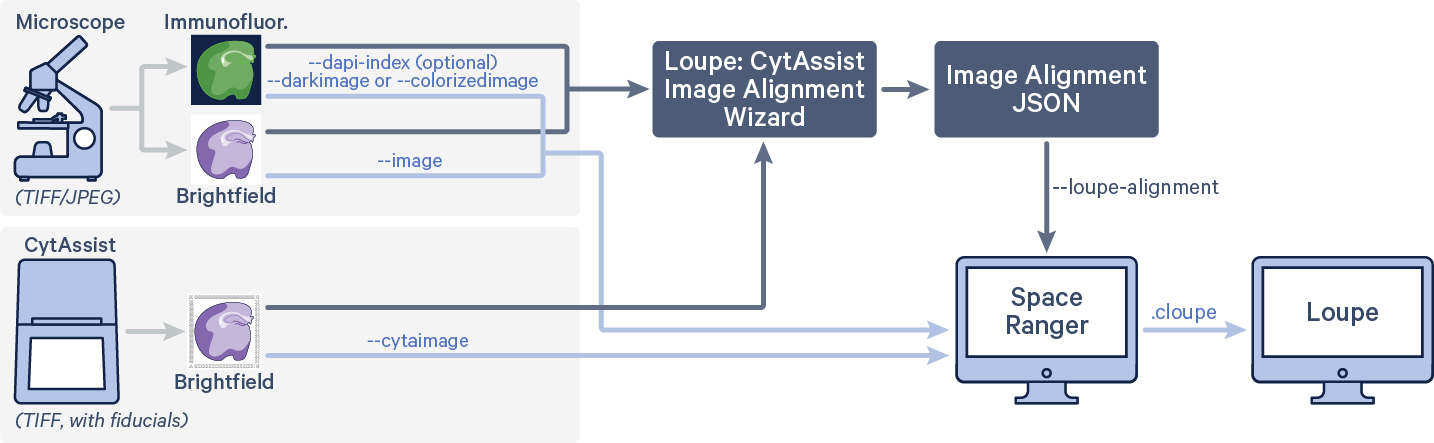
| Must use Space Ranger v2.0.1 or later, which contains a fix for the incorrectly thrown error in the Manual CytAssist Image Alignment workflow. See 2.0.1 release notes for details. |
If the automatic image registration between the CytAssist and microscope images is not satisfactory, use Loupe's CytAssist Image Alignment worflow to generate a image alignment JSON file. Both of the images used for alignment should also be passed to spaceranger count along with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --probe-set=/home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv\ #Path to Probe Set --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --cytaimage=/home/jdoe/runs/images/A1_sample345.tiff \ #Path to CytAssist image input --image=/home/jdoe/runs/images/sample345.tiff \ #Path to brightfield image input --slide=V49J01-123 \ #Slide ID --area=A1 \ #Capture area --loupe-alignment=sample345.json \ #Manual image registration file --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmode
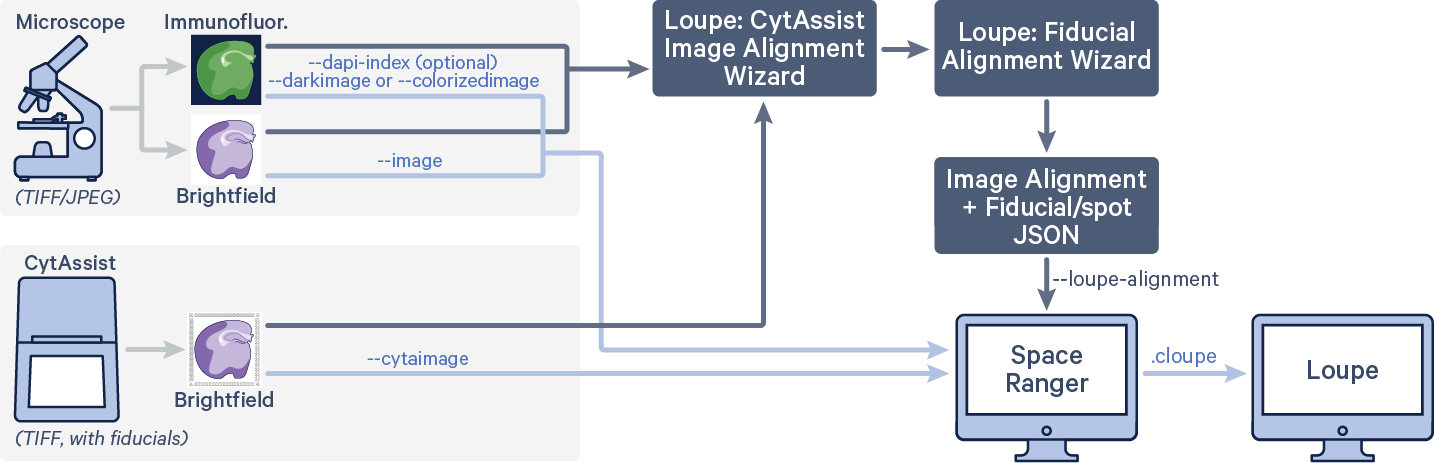
In some cases, the automatic image registration between the CytAssist and microscope images, or the automatic fiducial alignment and tissue detection on the CytAssist image, may not be satisfactory. In that scenario, both Loupe Browser's CytAssist Image Alignment and Manual Fiducial Alignment workflows can be used to generate a single JSON file that contains both image and fiducial alignment information. Note that both of the images used for alignment are passed to spaceranger count along with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --probe-set=/home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv\ #Path to Probe Set --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --cytaimage=/home/jdoe/runs/images/A1_sample345.tiff \ #Path to CytAssist image input --image=/home/jdoe/runs/images/sample345.tiff \ #Path to brightfield image input --slide=V49J01-123 \ #Slide ID --area=A1 \ #Capture area --loupe-alignment=sample345.json \ #Manual image registration and fiducial alignment file --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmode
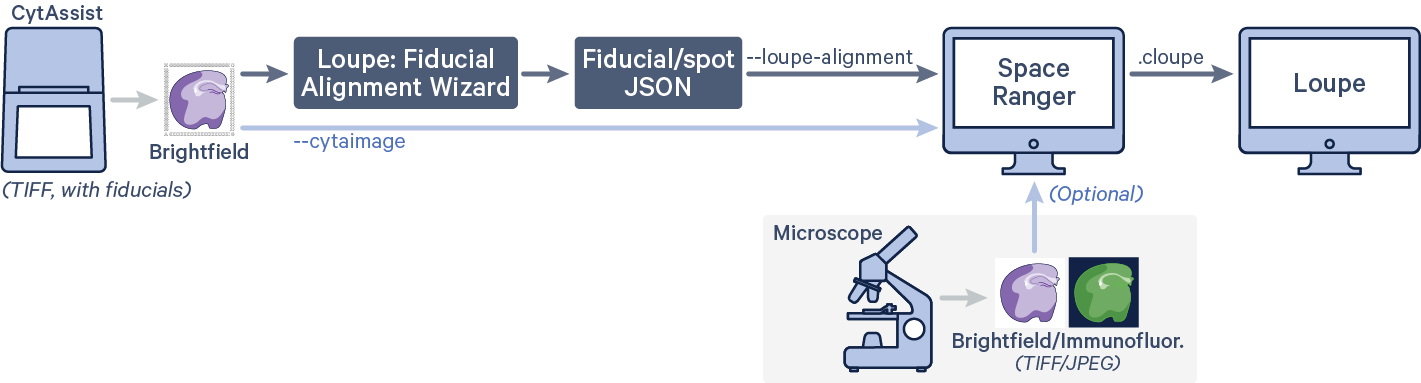
For CytAssist image-only input, there is also the option to run the manual fiducial
alignment workflow in Loupe Browser to generate a fiducial alignment and tissue assignment JSON file. Note that it is possible to pass a microscope image input in this configuration for automatic image registration in Space Ranger. The spaceranger count pipeline can be run with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --probe-set=/home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv\ #Path to Probe Set --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --cytaimage=/home/jdoe/runs/images/A1_sample345.tiff \ #Path to CytAssist image input --slide=V49J01-123 \ #Slide ID --area=A1 \ #Capture area --loupe-alignment=sample345.json \ #Manual fiducial alignment file --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmode
Probe set reference CSV file for human can be found in the probe_sets directory in the Space Ranger package. Note that only the highlighted probe set reference CSV files are compatible with CytAssist enabled Gene Expression workflow.
spaceranger-2.0.0/probe_sets/ ├── Visium_Human_Transcriptome_Probe_Set_v1.0_GRCh38-2020-A.csv ├── Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv └── Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv
The CSV files for both human and mouse
A successful spaceranger count run concludes with a message similar to this:
Outputs:
- Run summary HTML: /home/jdoe/runs/sample345/outs/web_summary.html
- Outputs of spatial pipeline:
aligned_fiducials: /home/jdoe/runs/sample345/outs/spatial/aligned_fiducials.jpg
detected_tissue_image: /home/jdoe/runs/sample345/outs/spatial/detected_tissue_image.jpg
scalefactors_json: /home/jdoe/runs/sample345/outs/spatial/scalefactors_json.json
tissue_hires_image: /home/jdoe/runs/sample345/outs/spatial/tissue_hires_image.png
tissue_lowres_image: /home/jdoe/runs/sample345/outs/spatial/tissue_lowres_image.png
cytassist_image: /home/jdoe/runs/sample345/outs/spatial/cytassist_image.tiff
aligned_tissue_image: /home/jdoe/runs/sample345/outs/spatial/aligned_tissue_image.jpg
tissue_positions: /home/jdoe/runs/sample345/outs/spatial/tissue_positions.csv
spatial_enrichment: /home/jdoe/runs/sample345/outs/spatial/spatial_enrichment.csv
barcode_fluorescence_intensity: /home/jdoe/runs/sample345/outs/spatial/barcode_fluorescence_intensity.csv
- Run summary CSV: /home/jdoe/runs/sample345/outs/metrics_summary.csv
- Correlation values between isotypes and Antibody features: null
- BAM: /home/jdoe/runs/sample345/outs/possorted_genome_bam.bam
- BAM BAI index: /home/jdoe/runs/sample345/outs/possorted_genome_bam.bam.bai
- BAM CSI index: null
- Filtered feature-barcode matrices MEX: /home/jdoe/runs/sample345/outs/filtered_feature_bc_matrix
- Filtered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/filtered_feature_bc_matrix.h5
- Unfiltered feature-barcode matrices MEX: /home/jdoe/runs/sample345/outs/raw_feature_bc_matrix
- Unfiltered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/raw_feature_bc_matrix.h5
- Secondary analysis output CSV: /home/jdoe/runs/sample345/outs/analysis
- Per-molecule read information: /home/jdoe/runs/sample345/outs/molecule_info.h5
- Loupe Browser file: /home/jdoe/runs/sample345/outs/cloupe.cloupe
- Feature Reference: null
- Target Panel file: null
- Probe Set file: /home/jdoe/runs/sample345/outs/probe_set.csv
Pipestance completed successfully!
The output of the pipeline is contained in a folder named with the sample ID you specified (such as sample345). If this folder already exists, in a rerun with the original parameters, spaceranger will assume it is an existing pipestance and attempt to resume running it. The subfolder named outs contains the main pipeline output files.
Once spaceranger count has successfully completed, you can further explore the results by
.cloupe file in
Loupe BrowserThe flags used in CytAssist enabled FFPE analysis are tabulated. For full list of all the relevant arguments refer to the Spatial GEX for FFPE page.
| Argument | Description |
|---|---|
--id | Required. A unique run ID string (e.g., sample345). The name is arbitrary and will be used to name the directory containing all pipeline-generated files and outputs. Only letters, numbers, underscores, and hyphens are allowed (maximum of 64 characters). |
--fastqs | Required. Either: Path of the fastq_path folder generated by spaceranger mkfastq (such as /home/jdoe/runs/HAWT7ADXX/outs/fastq_path). This contains a directory hierarchy that spaceranger count will automatically traverse.- OR - Any folder containing fastq files, for example if the fastq files were generated by a service provider and delivered outside the context of the mkfastq output directory structure. Can take multiple comma-separated paths, which is helpful if the same library was sequenced on multiple flowcells. Doing this will treat all reads from the library, across flowcells, as one sample. |
--sample | Optional. Sample name as specified in the sample sheet supplied to spaceranger mkfastq.
Can take multiple comma-separated values, which is helpful if the same library was sequenced on multiple flowcells and the sample name used (and therefore fastq file prefix) is not identical between them. Doing this will treat all reads from the library, across flowcells, as one sample. Allowable characters in sample names are letters, numbers, hyphens, and underscores. |
--probe-set | Required to enable FFPE analysis. Path to the probe set reference CSV file (such as /home/jdoe/spaceranger-2.0.0/probe_set/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv). |
--transcriptome | Required. Path to the Space Ranger compatible transcriptome reference (such as /home/jdoe/GRCh38-2020-A). |
--cytaimage | Required. CytAssist instrument captured eosin stained Brightfield tissue image with fiducial frame in TIFF format. The size of this image is set at 3k in both dimensions and this image should not be modified any way before passing it as input to either Space Ranger or Loupe Browser. |
--image | Optional for brightfield microscope image input. Brightfield tissue H&E image in JPEG or TIFF format. For more information on image file constraints, encoding and formats refer the Image Recommendations. |
--darkimage | Optional for dark background fluorescence microscope image input. Multi-channel, dark-background fluorescence image as either a single, multi-layer TIFF file or as multiple TIFF or JPEG files (which can provided as a comma separated list or by invoking the --darkimage parameter multiple times). Details on image file constraints, encoding and formats are described in the Image Recommendations. |
--dapi-index | Optional for dark background fluorescence microscope image input. The numeric value of the channel in a multi-channel fluorescence image corresponding to the tissue morphology counterstain such as DAPI. In absence of specifying this channel, the pipeline will use information from all the available channels. Note that channel numbers start from 1. |
--colorizedimage | Required for color composite fluorescence microscope image input. A color composite of one or more fluorescence image channels saved as a single-page, single-file color TIFF or JPEG. Please see the Image Recommendations for information on input image file formats. |
--slide | Visium slide serial number. Refer to the Slide Parameters for information on supported slide versions. Required unless --unknown-slide is passed. |
--area | Visium capture area identifier. Required unless --unknown-slide is passed. Options for standard Visium slides are A1, B1, C1, D1 and the options for XL Visium slides are A or B. |
--slidefile | Slide layout file indicating capture spot and fiducial spot positions. Required when no internet access unless --unknown-slide is passed. |
--unknown-slide | Optional. Set this if the slide serial number and area identifier are unknown. The value passed to this depends of the slide version. Possible values for CytAssist enabled workflow are visium-2 or visium-2-large. Setting this will cause Space Ranger to use default spot positions. Not compatible with --slide, --area, or --slidefile. Refer to the Slide Parameters for more information. |
--loupe-alignment | Required for manual alignment. Alignment file produced by the manual Loupe alignment step. A --image, --darkimage or --colorizedimage, the same image(s) that were used for manual alignment, must be supplied in this case. |
--filter-probes | Default:true. Include all non-deprecated probes listed in the probe set reference CSV file. Probes that are predicted to have off-target activity to homologous genes are excluded from analysis by default. Setting --filter-probes=false will result in UMI counts from all non-deprecated probes, including those with predicted off-target activity, to be used in the analysis. An alternative and preferable method to including all probes is described in this article. Probes whose ID is prefixed with DEPRECATED are always excluded from the analysis. |
--localcores | Recommended when run in localmode. Restricts spaceranger to use specified number of cores to execute pipeline stages. By default, spaceranger will use all of the cores available on your system. |
--localmem | Recommended when run in localmode. Restricts spaceranger to use specified amount of memory (in GB) to execute pipeline stages. By default, spaceranger will use 90% of the memory available on your system. |