Space Ranger2.0, printed on 03/26/2025
| This page lists the resources for analysis of CytAssist enabled whole transcriptome profiling. Refer to the corresponding pages for direct placement workflows for Fresh Frozen (FF) samples using poly-A based assay and formalin-fixed paraffin-embedded (FFPE) samples using probe-based assay. |
CytAssist instrument expands the capability of analyzing formalin-fixed paraffin-embedded (FFPE), Fresh Frozen (FF) or Fixed Frozen (FxF) samples to include tissue sections on archived standard glass slides. It is compatible with H&E and IF staining, adding the ability to preselect the best tissue section for analysis. By introducing slides with two different Capture Area sizes (6.5 mm or 11 mm), the CytAssist workflow offers flexibility in tissue size used for analysis.
The CytAssist instrument automates the transfer of molecules from the tissue section on a standard glass slide to the CytAssist Spatial Gene Expression Slide. The FFPE, FF or FxF tissue sections on standard glass slides follow the direct placement FFPE protocol until probe hybridization and ligation. CytAssist mediates the tissue permeabilization to release the ligation products for capture by spatially barcoded oligonucleotides within each Capture Area on the CytAssist Slide. The final product is a sequencing library constructed from the barcoded probes that is similar to a Visium for FFPE sequencing library.
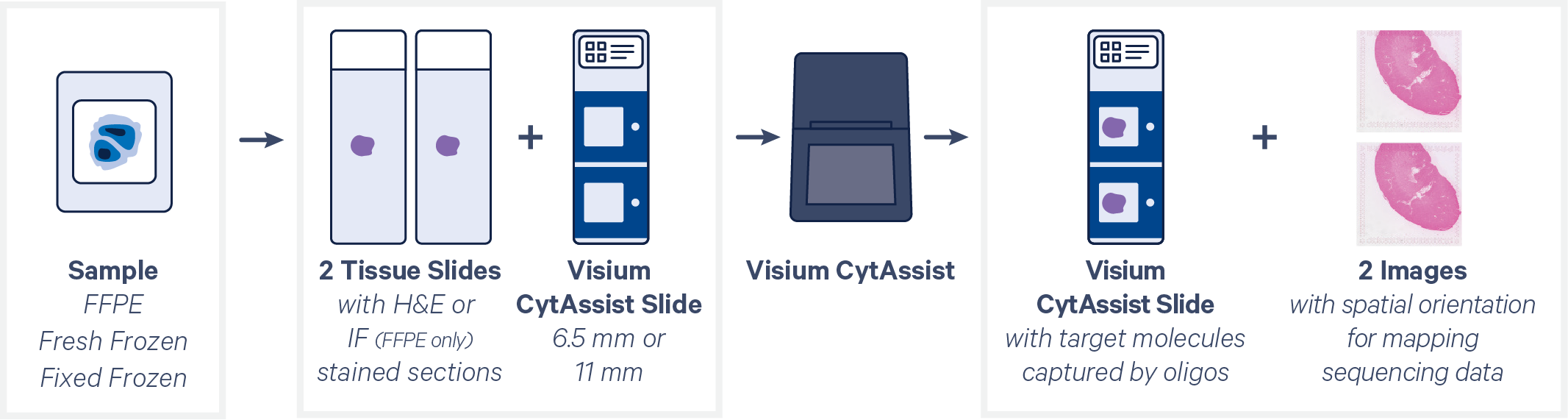
The analysis workflow for CytAssist mediated Spatial Gene Expression is similar to that for FFPE samples and uses a short read aligner tailored to probe sequences. To utilize the best imaging available for analysis, Space Ranger can take a brightfield or fluorescence microscopy image of the tissue section on the standard glass slide in addition to the CytAssist instrument generated image of the CytAssist slide. After image registration in Space Ranger, spatial gene expression information is mapped in the context of the high resolution tissue image. Refer to the imaging algorithms page for more details.
Resources and additional information are available below: