Space Ranger2.0, printed on 03/11/2025
| This page lists the resources for analysis of whole transcriptome profiling using Visium Spatial Gene Expression direct placement workflow in Fresh Frozen (FF) samples. Refer to the corresponding pages for direct placement workflow for formalin-fixed paraffin-embedded (FFPE) samples using probe-based assay and CytAssist enabled workflow for either FFPE or FF samples using probe-based assay. |
The Visium Spatial Gene Expression workflow measures RNA levels in intact, fresh frozen (FF) tissue sections, by capturing mRNA using spatially barcoded oligonucleotides attached to the Visium slide. The workflow begins by first determining the optimal permeabilization time for the tissue. The tissue is stained, and imaged followed by cell permeabilization to release mRNA which binds to the poly(dT) region of spatially barcoded oligonucleotides present on the slide surface. Libraries are generated from the barcoded cDNA of a captured transcript and are sequenced.
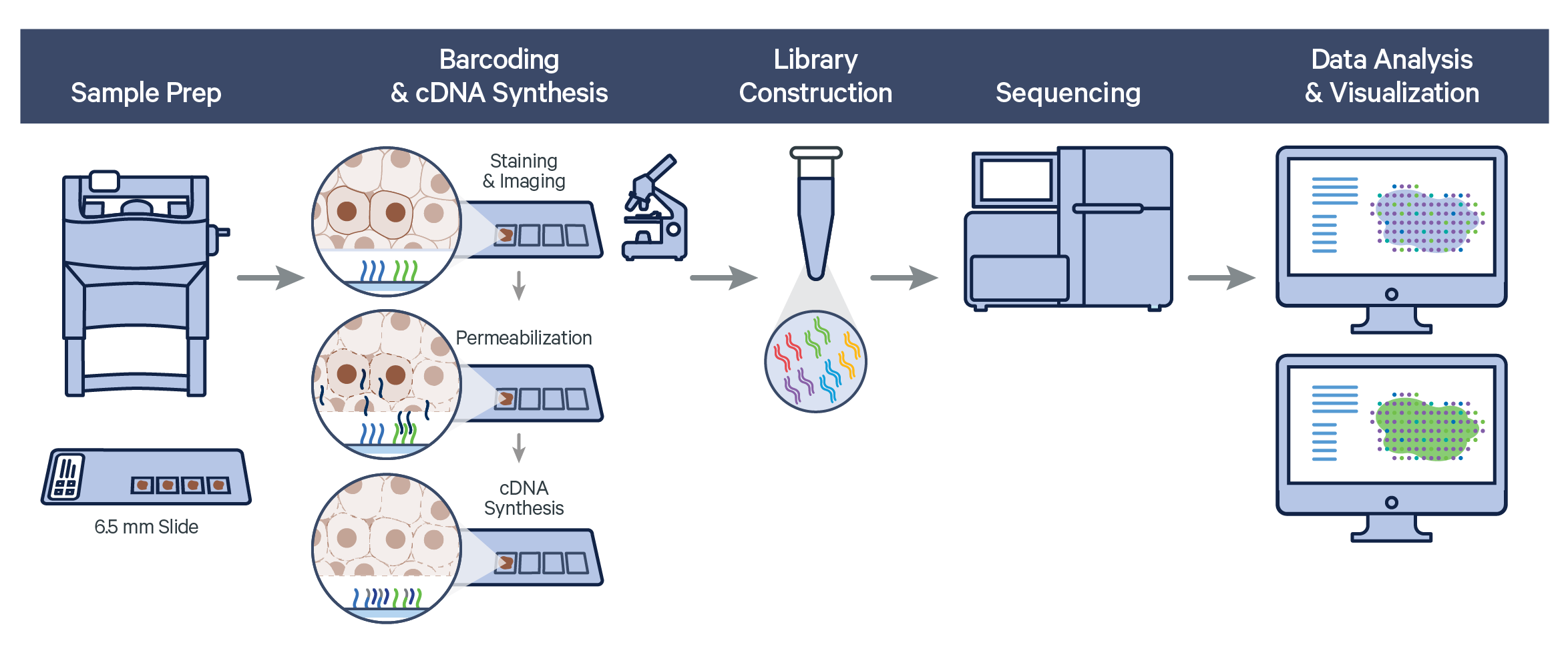
Visium Spatial Gene Expression relies on Space Ranger analysis software which uses STAR aligner to perform splicing-aware alignment of transcript reads to the genome. Space Ranger uses the transcript annotation GTF file to identify exonic reads, which when compatible with a single gene annotation, are considered for UMI counting.
Resources and additional information are available below: