Cell Ranger4.0, printed on 03/31/2025
Loupe V(D)J Browser includes a variety of tools to explore the diversity of clonotypes across your entire sample, and to more precisely hone in on individual clonotypes, genes, motifs, or even individual cell barcodes. You can use the summary charts to look at the diversity of your data at a high level, and use tools both within the clonotype list and the summary charts to filter clonotypes by a variety of criteria.
There are seven different charts through which you can investigate the diversity of a V(D)J sample: clonotype abundance, CDR3 abundance, V gene usage, D gene usage, J gene usage, C gene usage, and a V-J gene heatmap. Each chart comes with various viewing options and can be used to filter the clonotype list. They also support the ability to export the data as a CSV table, a PNG image, or an SVG drawing via the Export As button in the upper right. You can select a chart from the Chart Type dropdown at upper left. The question mark above the chart selector will show help text for the current chart, but here's an overview:
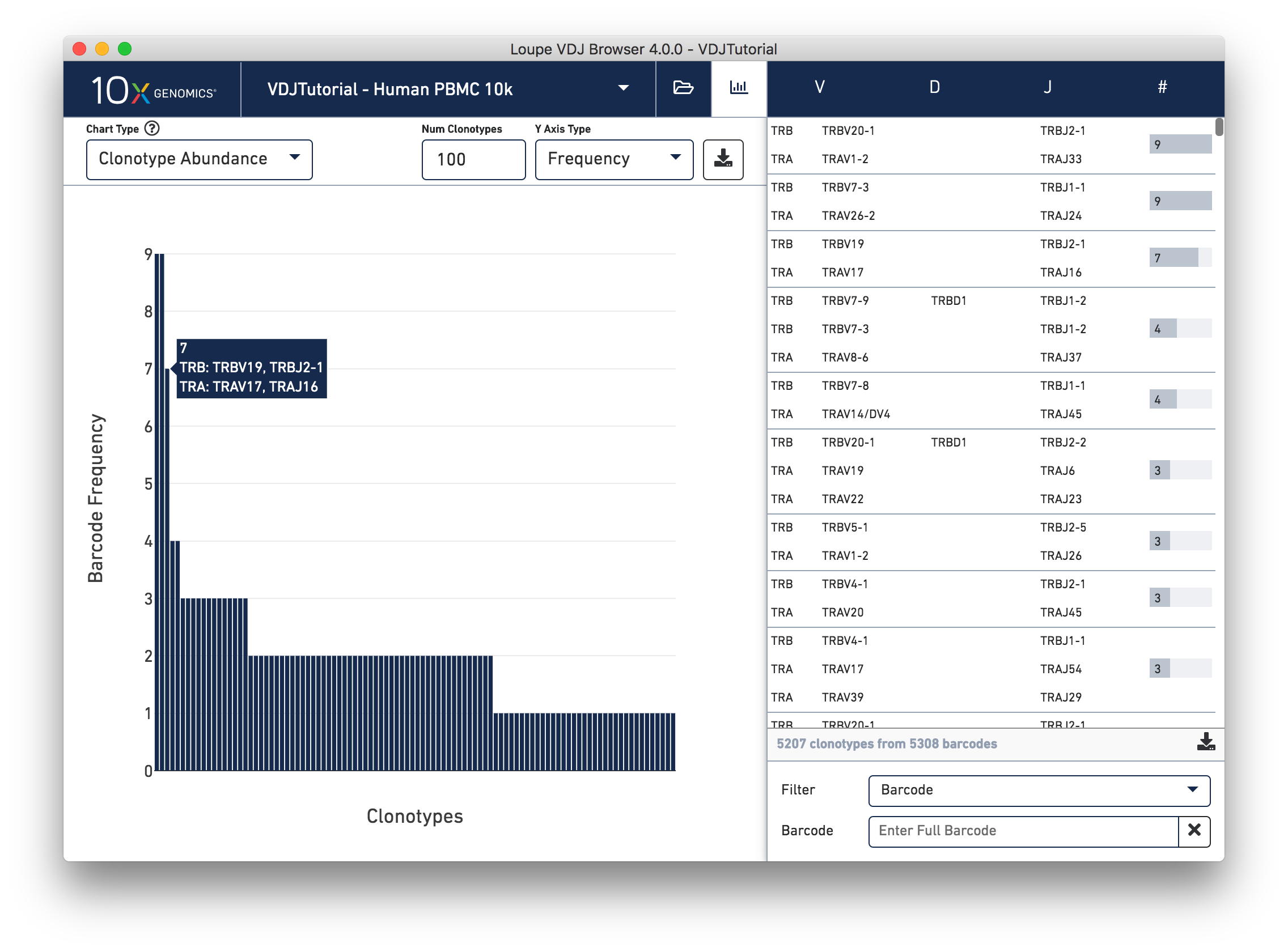
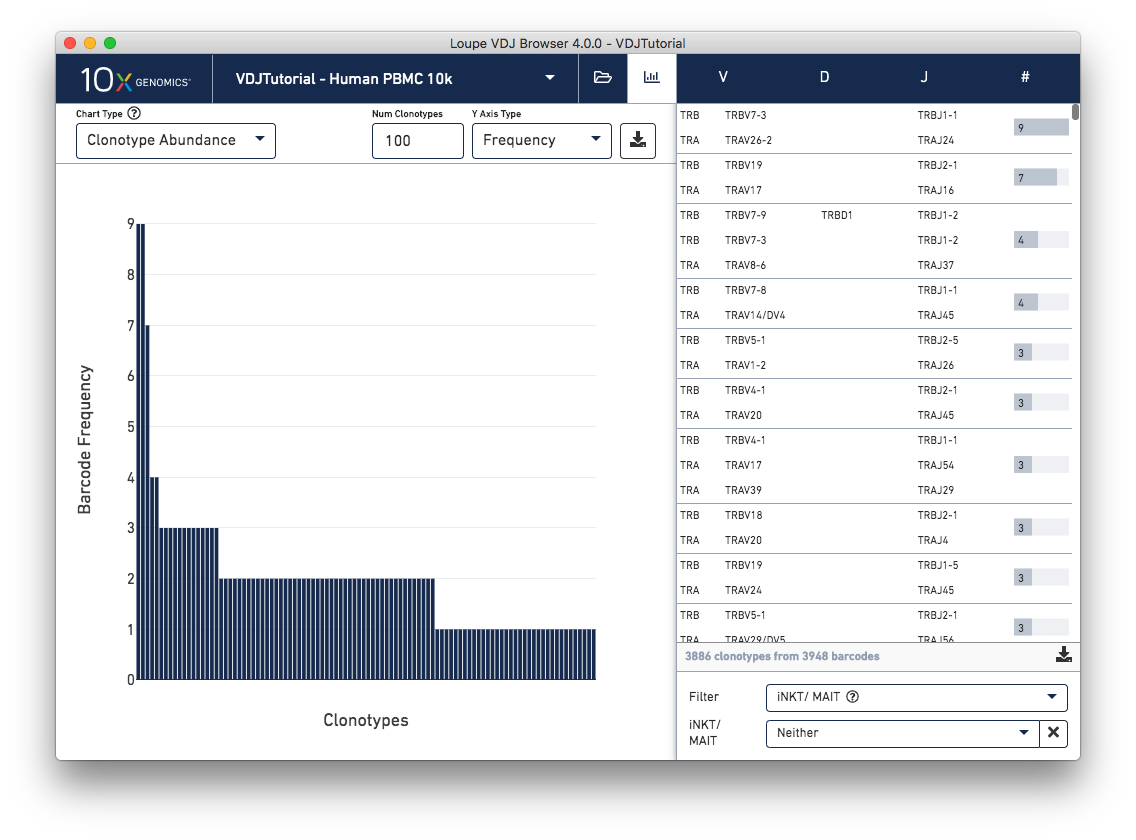
This chart shows the most frequent clonotypes, from most to least abundant. It's a more compact representation of the barcode counts in the clonotype list at right. You can change the number of top clonotypes to show by typing a new number in the Num Clonotypes input box above the chart. As with all bar charts in the Summary Chart View, you can change the y-axis to reflect either absolute clonotype count or percent of clonotypes (or genes) that each bar represents. You should see that the tutorial sample has three fairly abundant clonotypes, and a few other repeated clonotypes. Then abundance quickly drops off, and we see a long tail of singleton clonotypes represented once each.
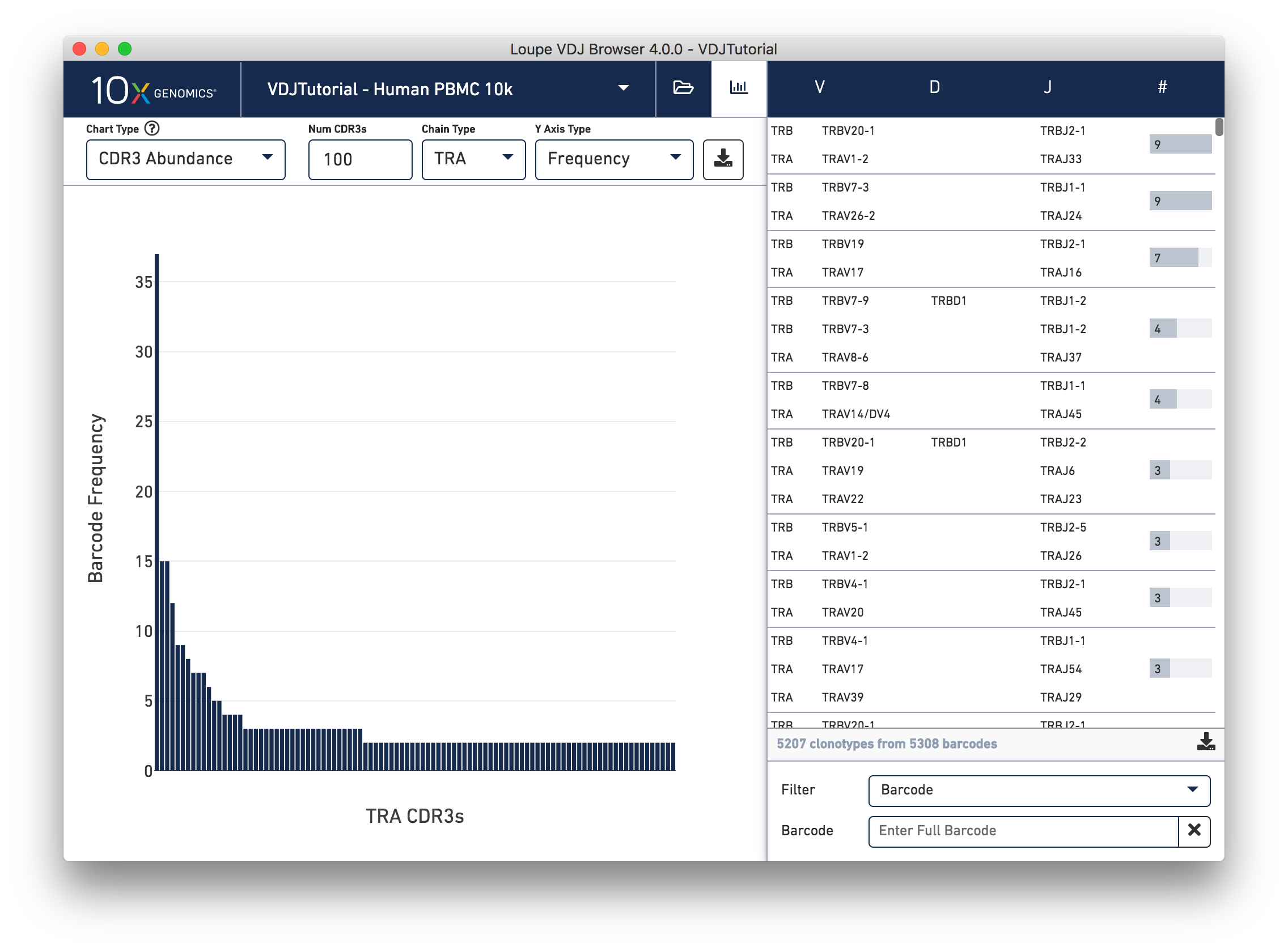
Select CDR3 Abundance from the Chart Type selector. This chart is similar to the Clonotype Abundance plot except that it shows the abundance of CDR3 sequences within individual chains. It counts all chains with each CDR3 detected in any cell in the sample. In this plot, you can view one chain type at a time.
Other features of the CDR3 Abundance chart are the same as the Clonotype Abundance plot. You can change the number of CDR3s shown, as well as the y-axis type.
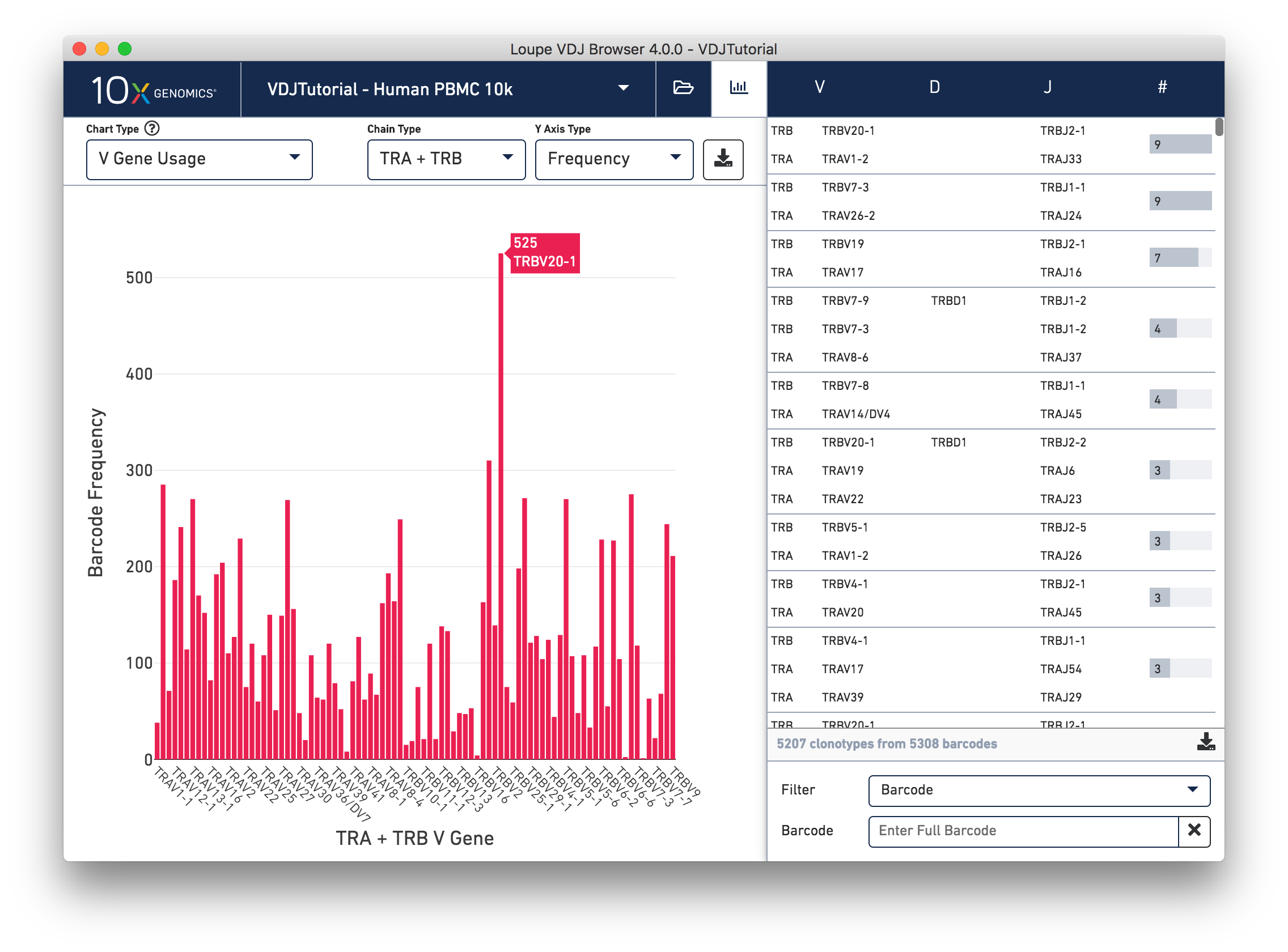
Select V Gene Usage from the Chart Type selector. The chart
in the content panel will change to a histogram of V gene frequency across the sample.
The y-axis is the number of cells in which a particular V gene was expressed.
You can see that the most numerous V gene in this sample is TRAV20-1,
which was detected in transcripts with 525 distinct cell barcodes.
The Chain Type selection box allows you to see all chain types or to limit the chart
to one chain type. In this case, we can see options TRA and TRB; if our sample included
B cells and not T cells, we would have IGH, IGK, and IGL instead.
The D, J, and C Gene Usage plots are analogous. Those charts show their respective gene usage distributions with the same functionality. You can view them by selecting them from the Chart Type selector.
Finally, select V-J Gene Heatmap from the Chart Type
selector to see the V-J Gene Heatmap. Through this chart, you can view the frequency of
V-J gene pairings across your sample. Each box in the heatmap represents the number
of cells in which Cell Ranger detected transcripts with the specified V gene (column)
and J gene (row). If a particular box is yellow, no such transcripts were detected in the sample.
Darker colors show more frequent V-J gene pairings. If you hover over each box, you'll see
the names of the V and J genes as well as the frequency of the pairing. After hovering
the TRAV26-1/TRAJ34 cell, you should see the cell count as shown below:
You can use the tools below the clonotype list to find clonotypes in a variety of ways. You can select a filtering method via the Filter By box, and enter a value in the input box to its right. To clear a filter and see the complete clonotype list, press the X button at bottom right. Each filtering method is a little different, as described below:
When the Gene Name filtering method is selected, Loupe V(D)J Browser will attempt to autocomplete the gene you are looking for by popping up a menu above the input box as you start typing. You can select a gene with your mouse or press enter. Selecting a gene will add it to the filter list. The clonotype list will only show clonotypes that contain at least one chain that match all the gene annotations in the input box. For example, searching a TRAV gene and TRBJ gene together should yield no results. To remove a gene, you can press the X next to a gene, or press delete.
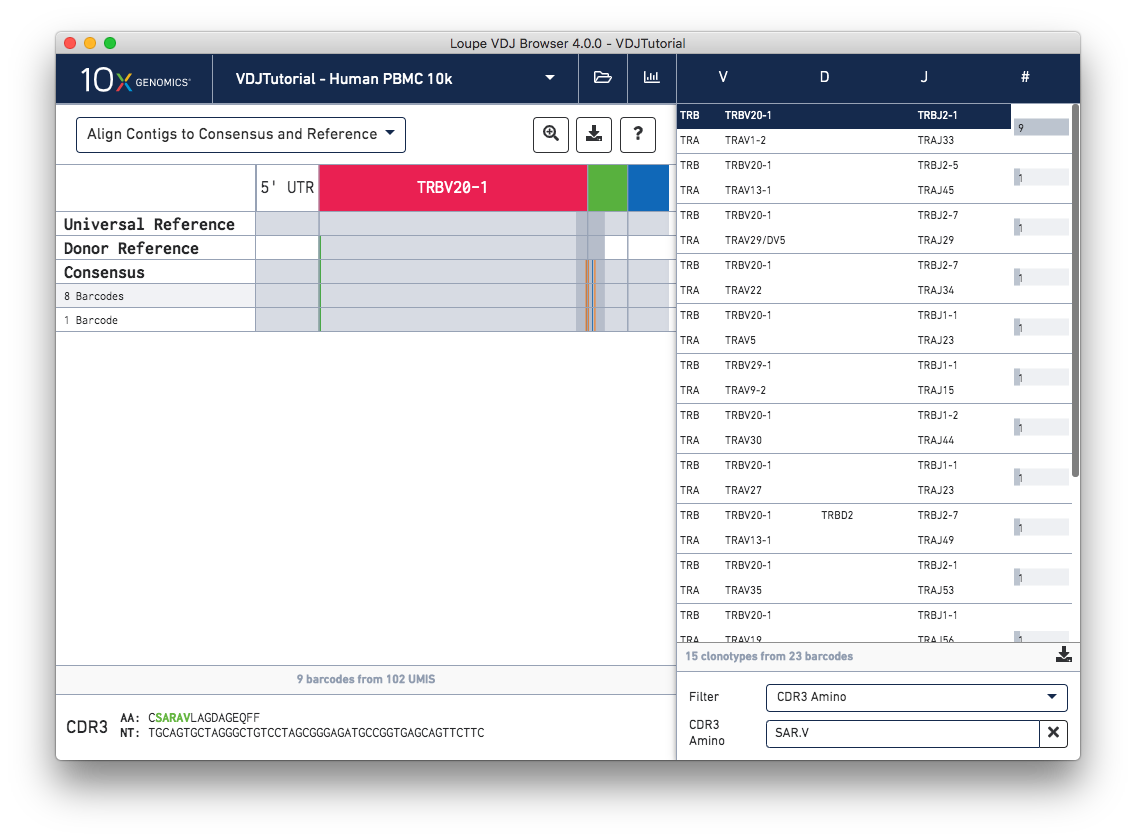
You can filter by CDR3 Amino (residues), or by CDR3 Bases (nucleotides). In both cases,
the clonotype list will be filtered by partial CDR3 match; that is, if the CDR3 of any
chain in a clonotype contains those residues or bases, the clonotype will remain in the list.
Use the '.' character to include a wild-card residue or base match in both modes. For
example, in CDR3 Amino mode, the SAR.V search sequence will find chains with
SARAV, SARIV, SARPV residues, among others.
For T cell datasets identifies clonotypes with potential invariant natural killer T (iNKT) and mucosal-associated invariant T (MAIT) cells using the iNKT/MAIT Evidence filter. The resulting list will show clonotypes with cells that have supporting evidence in any of the TRA genes, TRA junctions, TRB genes, or TRB junctions of being an iNKT or MAIT cell. Further details on the identification criteria can be found in the Cell Ranger documentation. In addition to identifying clonotypes with iNKT evidence or MAIT evidence, there is also the option to filter out clonotypes with any evidence using the Neither filter, or use Both for results that show iNKT and MAIT evidence in the same clonotype.
If you have a matching 5′ gene expression dataset, you can load the .cloupe file
generated by cellranger count or cellranger multi into the Loupe V(D)J Browser to
filter the clonotype list by clusters created in Loupe Browser. To learn how to do
this as well as other analyses with matching 5′ gene expression and V(D)J data,
check out the Loupe Integrated Gene Expression and V(D)J Analysis Tutorial.
You may want to search for individual cell barcodes identified by secondary analysis, and searching by barcode allows you to pinpoint these individual cells. The barcode must be an exact match; there are no wild cards allowed.
After gaining the ability to identify clonotypes of interest, it's time to explore them in more detail, by exploring individual chains.