Cell Ranger, printed on 04/05/2025
Loupe Browser 5.0 has been updated to allow users to filter and recluster single-cell gene expression datasets. With Loupe's new intuitive, high-performance filtering and reclustering workflow, users can filter out low-quality cell barcodes, narrow on cell types of interest for a more focused study, and explore and share the results of their reanalysis, all interactively, without writing a single line of code.
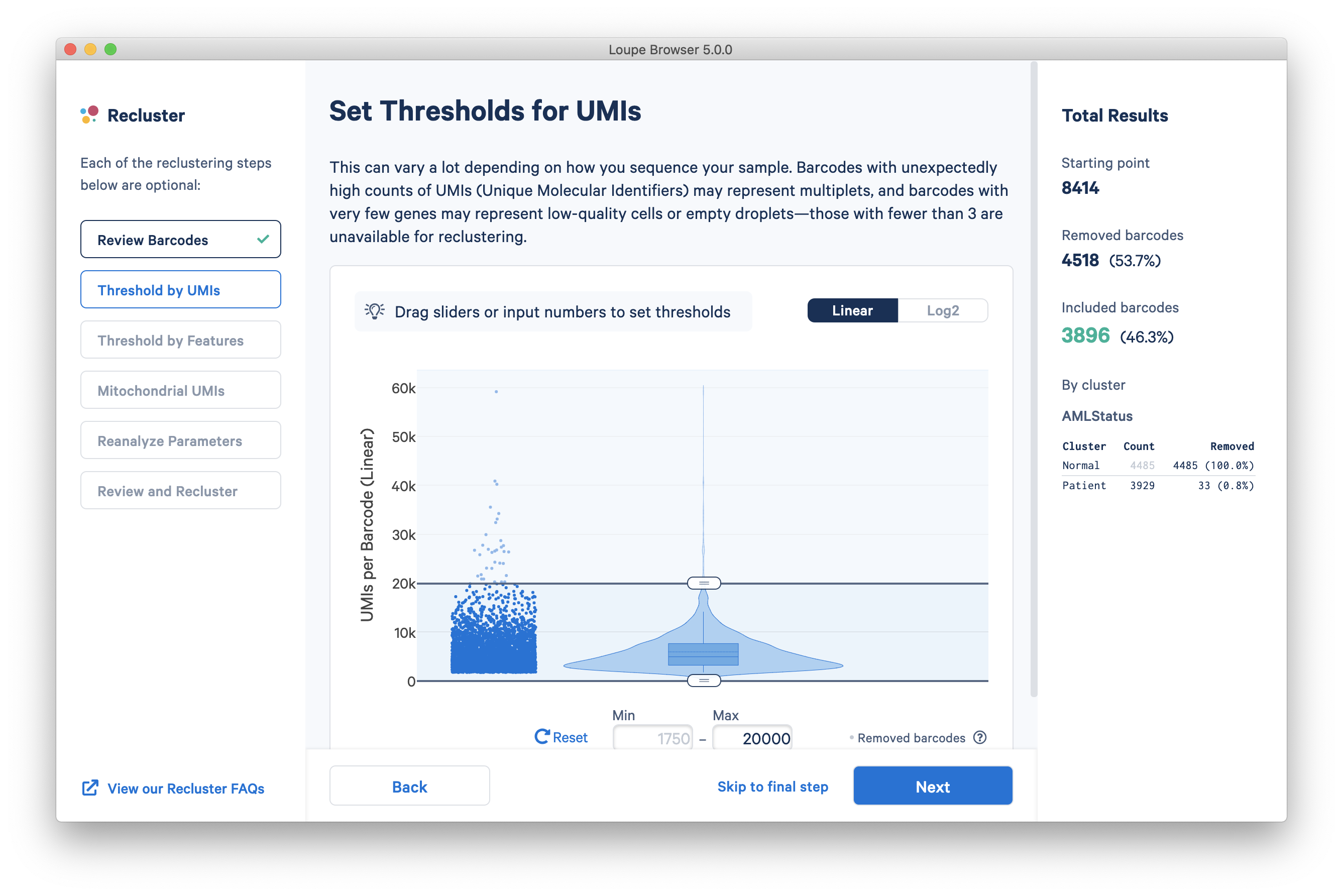
The reclustering workflow allows for inclusion or exclusion of barcodes by:
The reclustering algorithm generates a new Louvain-based clustering and a t-SNE plot over the filtered dataset. Datasets with less than 10,000 cells should be processed in under two minutes, and Loupe supports reclustering operations on datasets as large as 100,000 cells.
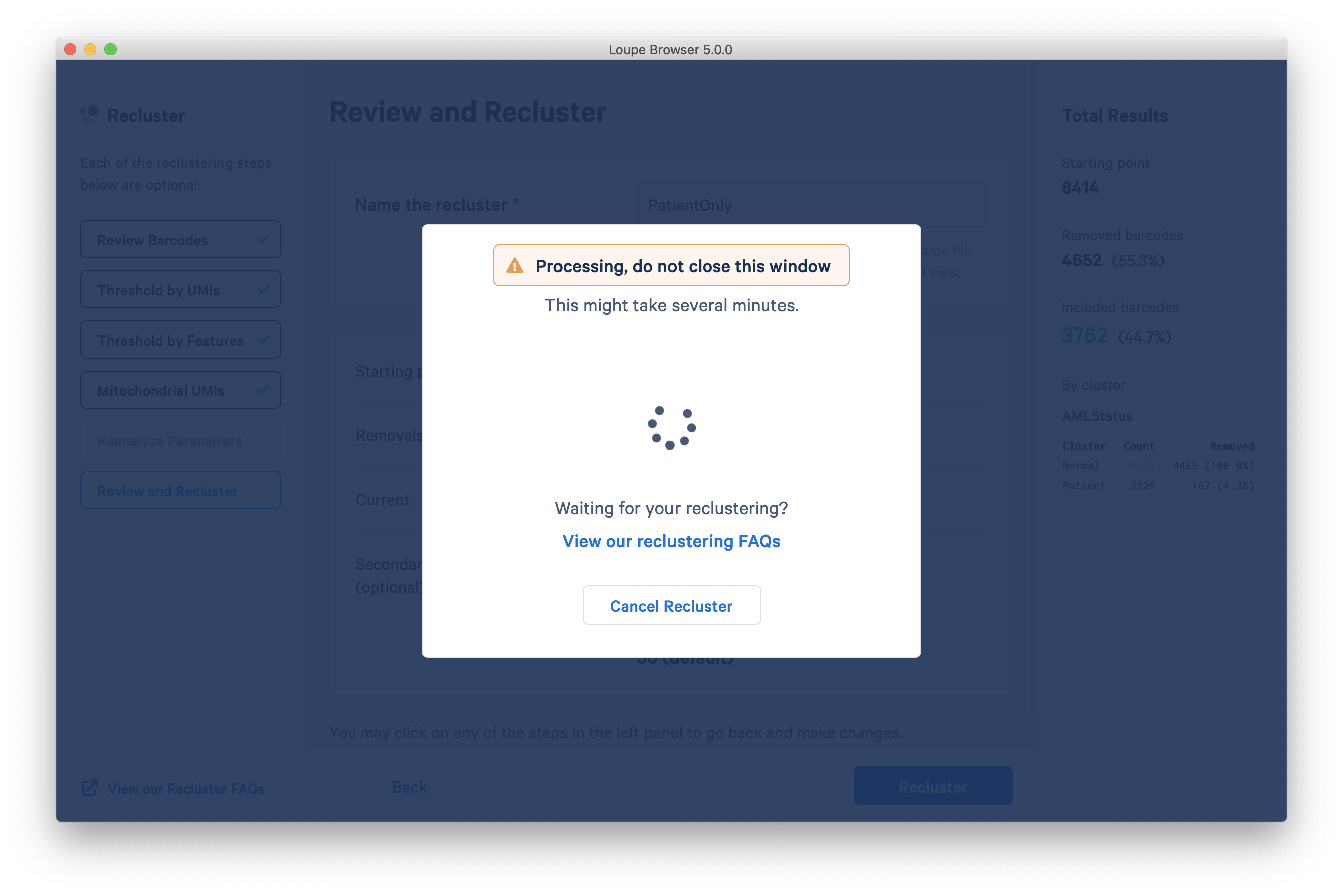
For more information and a guide to the workflow, visit the filtering and reclustering page.
Loupe Browser 5.0 includes updates to the clonotype table, with new filtering options
and improved performance with high cell count .vloupe files.
Loupe Browser 5.0 now only supports the loading of VDJ clonotypes created with Cell Ranger 5.0 and later. Previous versions of Loupe Browser may be used to look at VDJ data generated by Cell Ranger 4.0 and earlier.
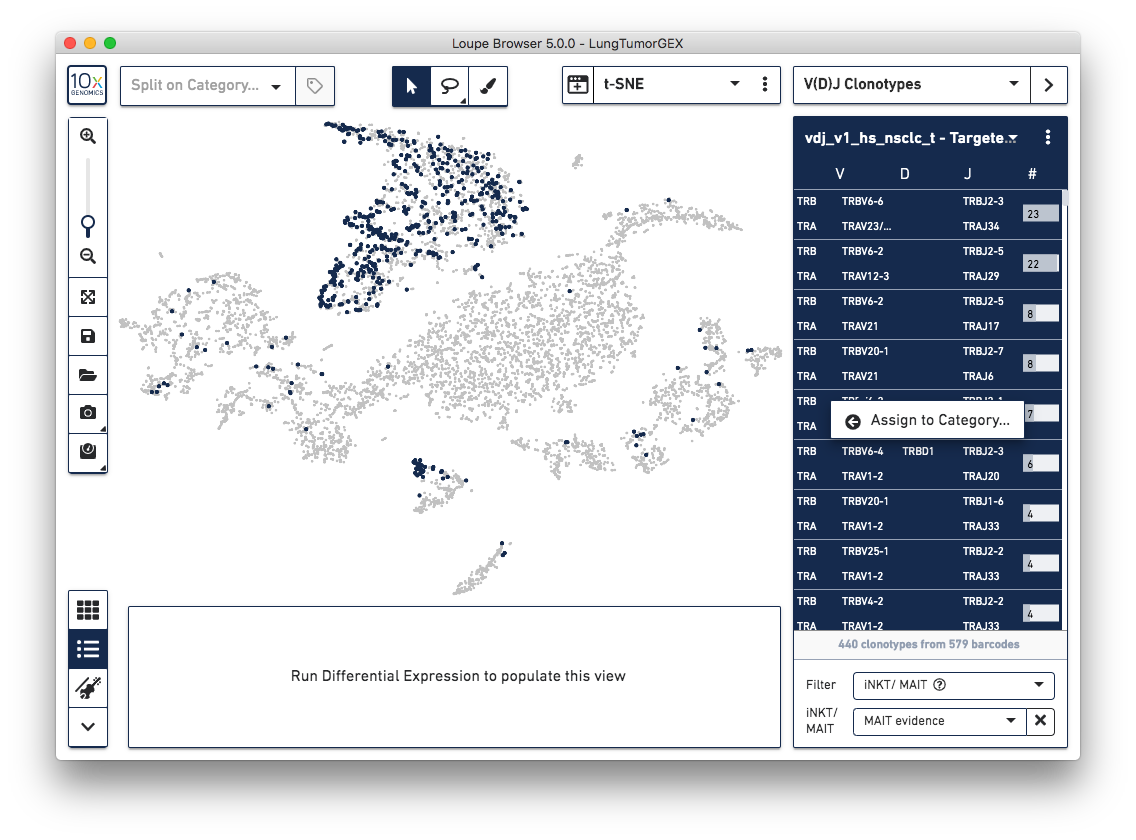
Identify potential invariant natural killer T (iNKT) cells and mucosal-associated invariant T (MAIT) cells with a new clonotype table filter. The iNKT/MAIT evidence filter can be used to view clonotypes that contain any cells with supporting evidence in any of the TRA genes, TRA junctions, TRB genes, or TRB junctions of being an iNKT or MAIT cell. This filter is only visible for T cell datasets.
There are other powerful filtering options added in this release:
.vloupe files generated by
aggr.Loupe Browser 5.0 can be installed concurrently with a currently installed version of Loupe Browser 4.0 or lower. This is required to be able to support visualization of VDJ data created by Cell Ranger 4.0 or earlier.
Differential expression is now more intuitively tied to the active category. If a differential expression calculation has not been run for the currently selected category, the differential expression table and heatmap will appear blank and prompt you to run the calcuation, instead of showing the most recently calculated differential expression.