Cell Ranger4.0, printed on 04/03/2025
To learn how to navigate the Loupe Browser interface, a pre-loaded AML tutorial dataset is included and used to demonstrate the interactive functionality.
View Panel
The workspace is centered around the view panel, in which single points representing cell barcodes are shown in a variety of projections. Each point represents a single barcode, the vast majority of which correspond to a single cell. The default projection is the t-SNE plot created by the Cell Ranger pipeline. Cell Ranger generates this plot by identifying the most significant feature vectors using principal component analysis, and then applying t-SNE dimensionality reduction to produce a two-dimensional scatter plot. A selector at the upper right of the barcode plot allows you to toggle between projections, including UMAP. For datasets generated using Feature Barcode technology, you may view alternate t-SNE or UMAP plots generated by considering cell surface markers or CRISPR guide RNA features instead of genes. You may also switch to a projection that plots feature counts on two-dimensional axes.
You can drag the mouse over the cells to reposition the plot, and use the mouse wheel or track pad to smoothly zoom in and out. You'll see cluster labels as you move your mouse over the plot, which is useful for data that has a high number of precomputed clusters. Cells are colored by the active legend in the sidebar.
Note: UMAP projections are only generated by Cell Ranger 3.1 and later.
The following image provides an overview of the key components of the Loupe Browser interface. Each of these components are described in more detail below.
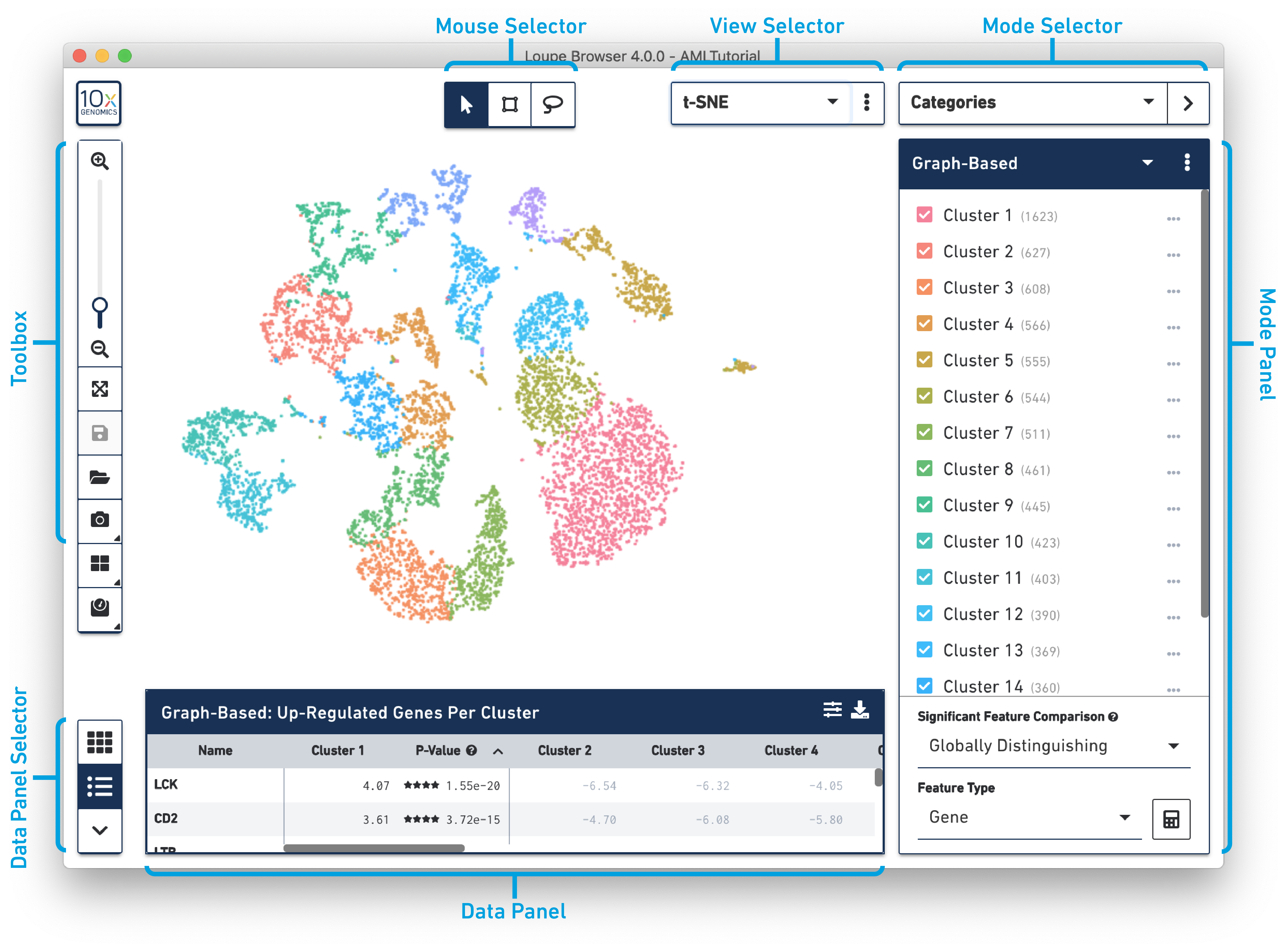
The Mouse Selector has three options from left to right:
| Tool | Description |
|---|---|
 |
Pan - Move the image up and down, or left and right. |
 |
Rectangular Selection - Select and label a rectangular image area. |
 |
Lasso Selection - Select and label an image area with an irregular shape. |
The Toolbox is on the left side of the window. Move your mouse over the toolbox icons to see an explanation of what each tool does. The tools in the toolbox perform the functions listed in the table below.
| Tool | Description |
|---|---|
 |
Zoom In and Zoom Out Click on + to Zoom In, click on - to Zoom Out, or use the slide bar to zoom in and out. |
 |
Autoscale Autoscale to fit the screen after zooming in or out. |
 |
Save As After changes have been made to a .cloupe file, click Save As, name the file, and select a location for the file. |
 |
Open File Open a new .cloupe file |
 |
Export Plot Click on the triangle in the bottom right corner and select Export Screenshot as PNG. |
 |
Split View Split View can be used with t-SNE, UMAP and Feature Plot to view clusters displayed in the Mode Panel. |
 |
Marker Settings Marker Settings can be used with t-SNE, UMAP and Feature Plot to scale the image automatically or by percentage. Uncheck the Auto-scale box to choose a percentage. |
To return to the home screen and the Recent Files list, click the 10x Genomics button in the top left corner.
The View Selector controls what is displayed in the View Panel. There are three different types of views that can be selected for gene expression datasets:
The following components in the interface are common across all views and are defined in separate sections:
Loupe Browser provides two different projection views of the data: t-SNE and UMAP. In these views, the cells are displayed based on either the t-SNE or UMAP algorithms in Space Ranger. Each cell is color-coded based on the active legend in the Mode Panel.
In both of the projection views, dragging the mouse over the display area repositions the projection, and the mouse wheel or track pad can be used to smoothly zoom in and out. The cluster label associated with a cell is shown when you hover your mouse over that cell.
Please note that UMAP projections are only generated by Cell Ranger 3.1 and later.
The Feature Plot view allows you to visualize the expression levels of one or two genes for each cell. This view makes it easy to threshold sets of cells based on the level of expression of one or two genes. Features, in this case genes, can be entered in the text box at the top of the Y axis or on the right side of the X axis. These selectors also contain a control to switch the scale of the axis between linear and log scale.
There are many scripts and third-party tools that can be used to generate different projections of the data including but not limited to the following:
If you generate alternate projection coordinates from customer analysis, you can input that projection into Loupe Browser. To do this, click on the three vertical dots in the View Selector to import a csv file to use a custom projection for a dataset.
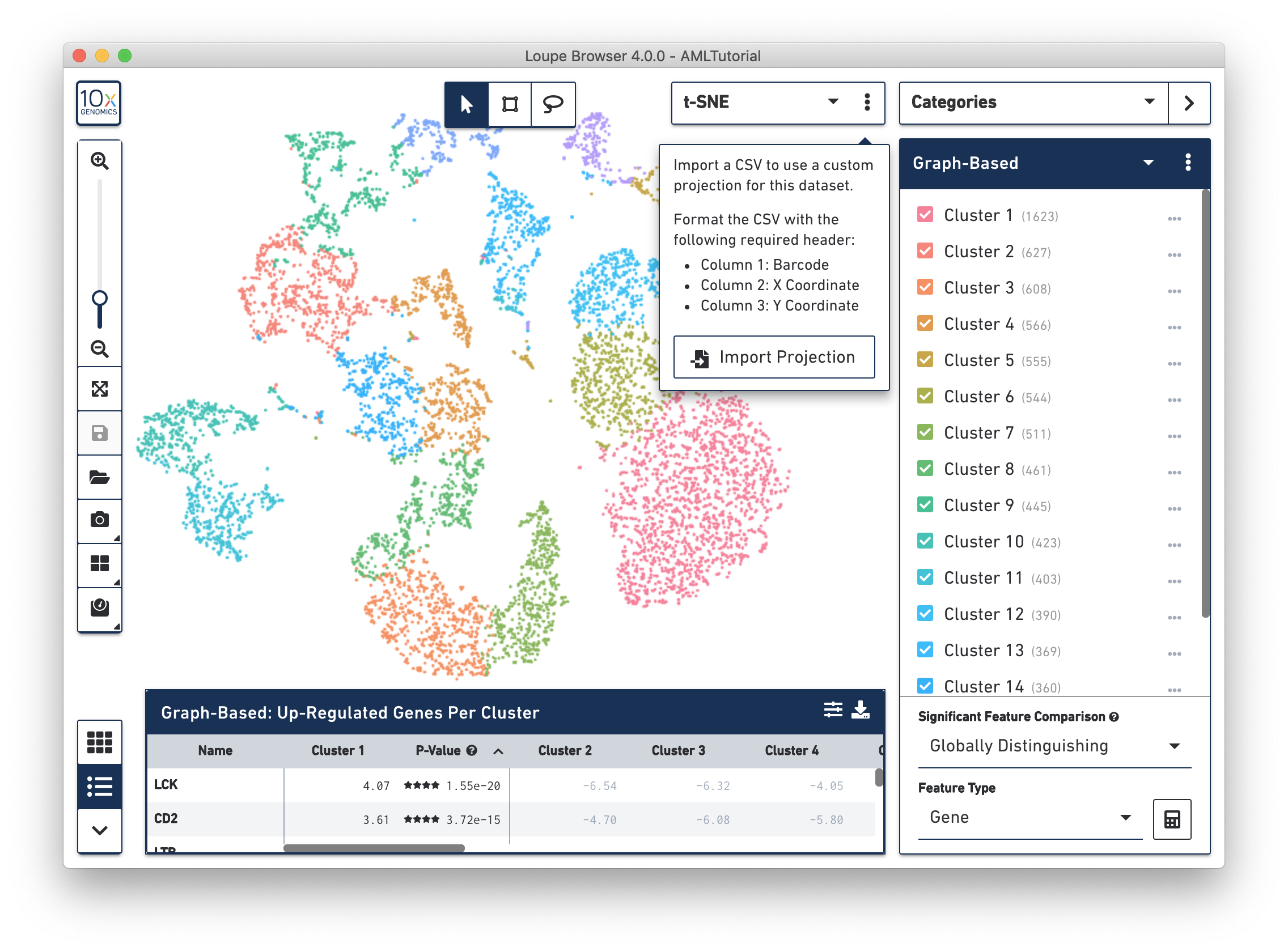
The csv format for importing projections is as follows:
To import new projections, click on Import Projection, select the csv file, and click on Import Projection. Once a custom projection is uploaded, click on Import Projection again to edit the name of the projection, or to delete it.
Use the mode selector in the top right corner of the workspace to switch between Loupe Browser's different modes. Switching between modes applies mode-specific coloring to the View Panel, and changes mode-specific functionality to the Mode Panel. There are four modes in Loupe Browser:
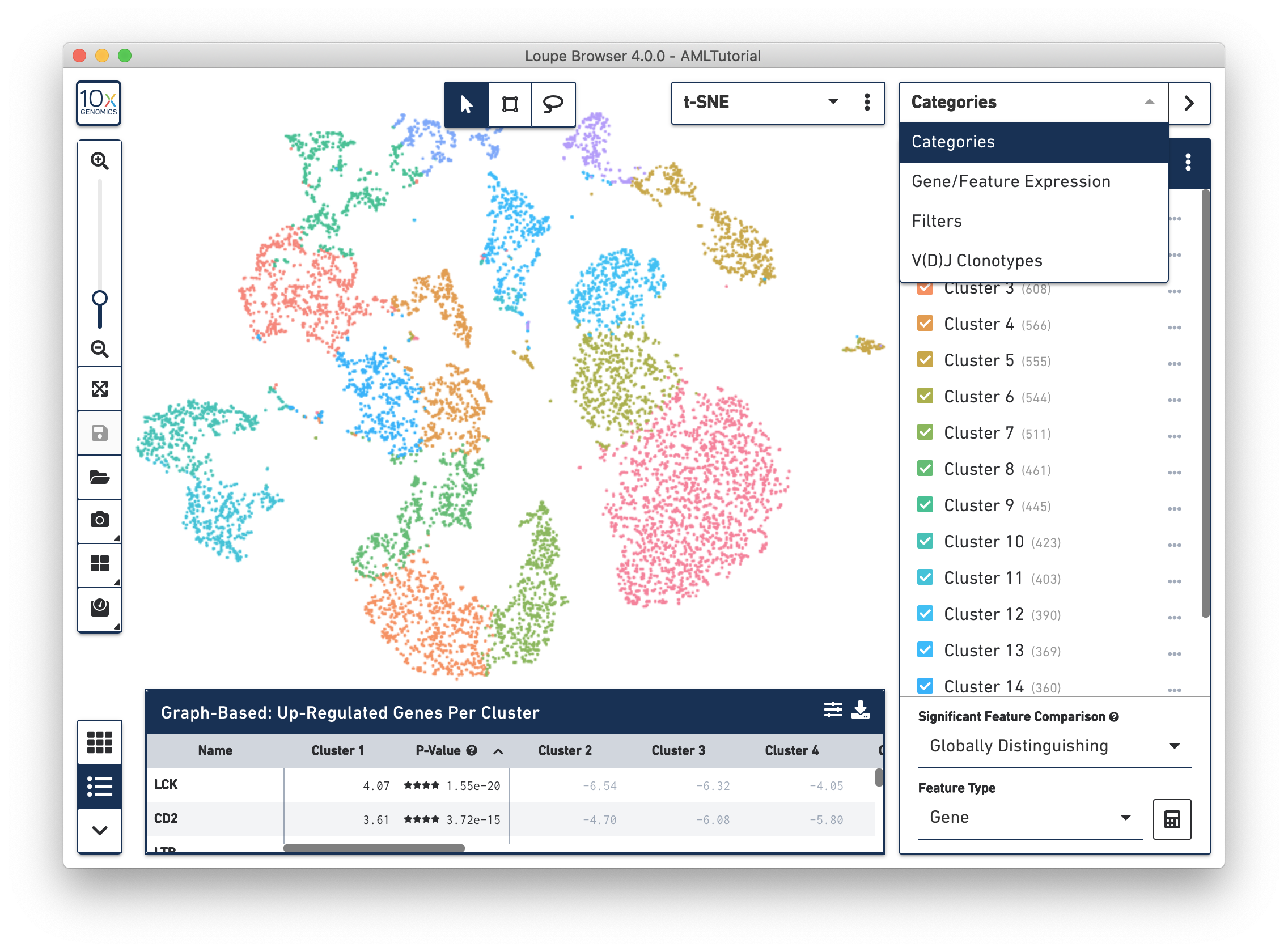
Categories Mode provides three ways to display data clusters, including:
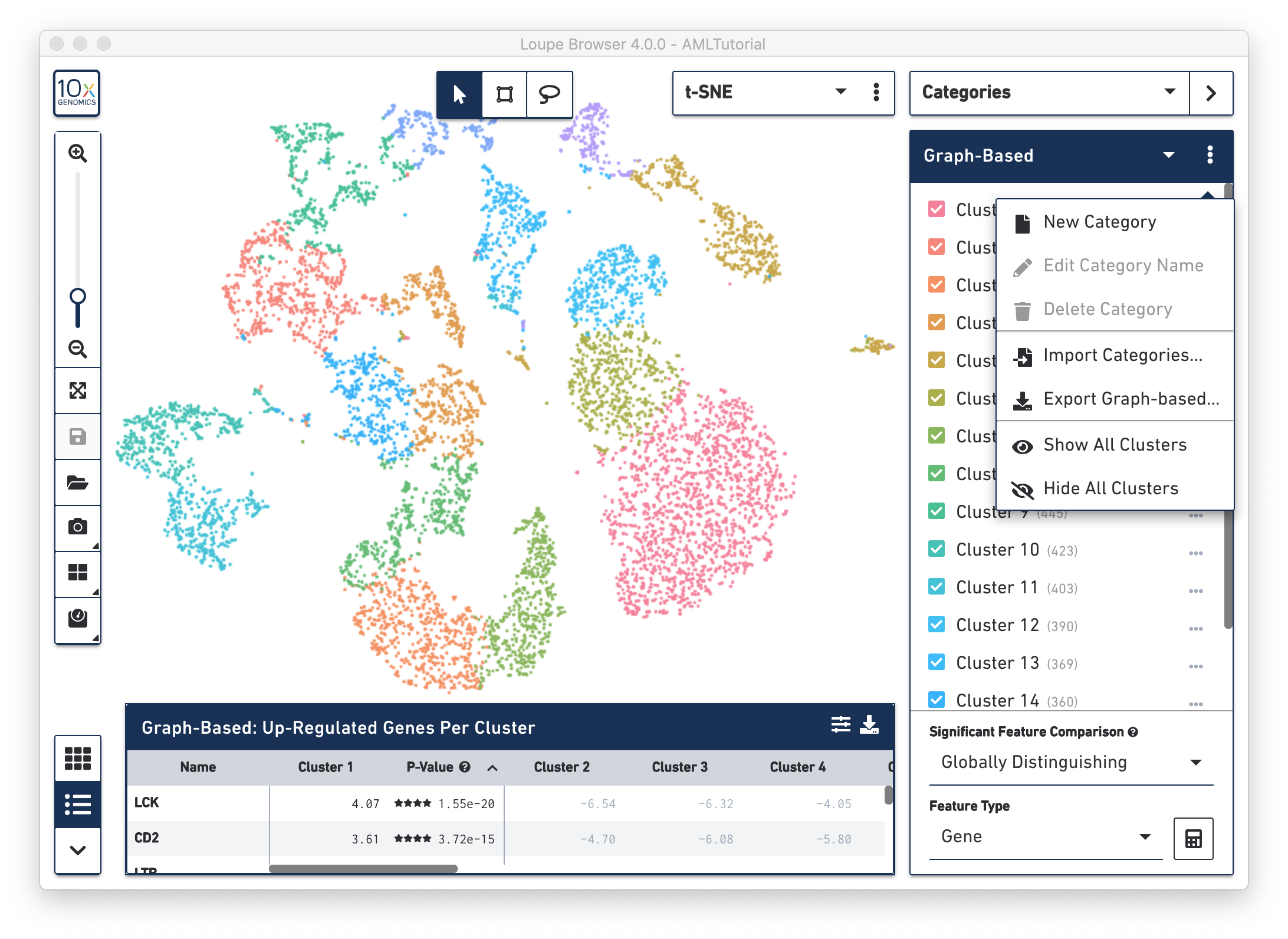
When you are in Categories Mode, you can control the content in the Mode Panel by clicking on the three vertical dots next to the drop-down menu.
Use this option to create a custom category and associated custom clusters.
This option is only applicable to custom categories.
This option is only applicable to custom categories.
Use this option to upload a csv file that contains customer cluster definitions on a per-barcode basis. The format of the csv file is as follows:
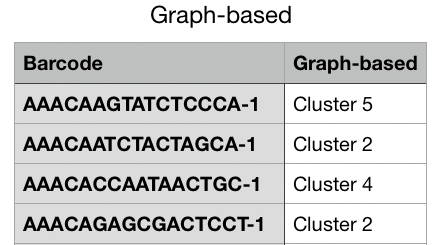
For this option, the [Name of Category] reflects the name of the current Category that you are viewing. This option exports a complete list of barcodes for the current Category and the associated cluster name that is associated with that Category. For example, if you are currently viewing the Graph-Based category, then this option in the drop-down menu is Export Graph-based. When you select this option, you can download a csv file that contains a list of all barcodes in the first column and their associated graph-based cluster label in the second column. See the example below.
This option automatically selects all clusters in the Mode Panel.
This option automatically deselects all clusters in the Mode Panel.
The Significant Feature Comparison functionality determines what features in the
dataset distinguish one or more clusters, and allows differential expression
analysis between clusters. There are two types of controls for this
functionality. Select options for these two controls using the drop-down menus
in the bottom right-hand corner. Once options are selected, click on the
![]() icon to execute the comparative analysis.
icon to execute the comparative analysis.
There are two different methods available for conducting a Significant Feature Comparison analysis.
Use this drop-down menu to select which type of feature you want to use in the comparison. The options are Gene or a specific Feature Barcode type, such as Antibody or CRISPR.
The Gene/Feature Expression mode provides a graphical representation of gene or feature expression across a dataset. One or more features can be viewed at a time. You can search for a feature of interest, or upload and save lists of features.
The Active Feature List contains a list of the features that you have either searched for, or uploaded. The expression patterns of those features are displayed in the View Panel. If more than one feature is in the Active Feature List, then the expression pattern corresponds to a combination of all the features. If only one feature is present or selected in the Active Feature List, then the expression pattern corresponds to that feature.
At the bottom of the Active Feature List there are a number of options that control how the data is visualized in the View Panel. The Scale & Attribute parameters control how the expression patterns are rendered in the View Panel. The top left menu sets which scale value to display. The top right menu sets how to combine values when there are multiple features in the Active Feature List.
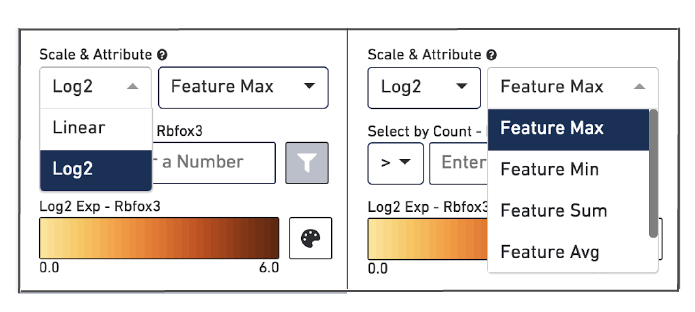
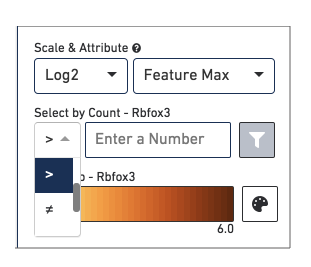
The Select by Count controls how to filter the expression values displayed.
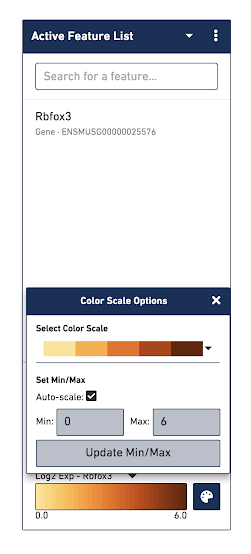
The color palette in the bottom right controls the color scale and range of values.You can also choose to manually set the min and max of the color scale by unchecking the Auto-scale checkbox, typing in a value, and clicking the Update Min/Max button. When setting manual min and max values, barcodes with values outside the range, less than the minimum or greater than the maximum, are colored gray. This is particularly useful if there is a lot of noise or ambient expression of a gene. Increasing the minimum value of the scale filters that noise. It is also useful to configure the scale to optimally highlight the expression of genes of interest.
In Gene/Feature Expression Mode, you can control the content in the Mode Panel under the Active Feature List by clicking on the three vertical dots next to the drop-down menu. This gives a number of options, as described below.
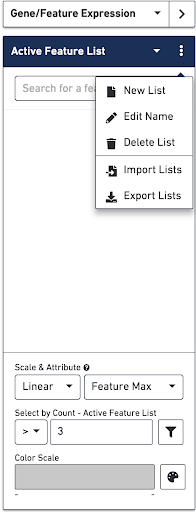
Use this option to create a custom list of features to visualize.
Edit the name of a list.
Delete a list.
Use this option to upload a csv file that contains a custom set of markers that results in one or more new lists. The format of the csv file is as follows:
If you have manually added features to a list, use this option to export the list of features as a csv file. This is useful if you want to reuse a list for other samples visualized in Loupe.
In Filter mode, you can compose complex boolean filters to find barcodes which fulfill your criteria. You can create rules based on feature counts or cluster membership and combine these rules using boolean operators. You can then save and load filters and use them across multiple datasets. For more details on using the Filter Mode, see the following tutorial built around Single Cell Gene Expression data.
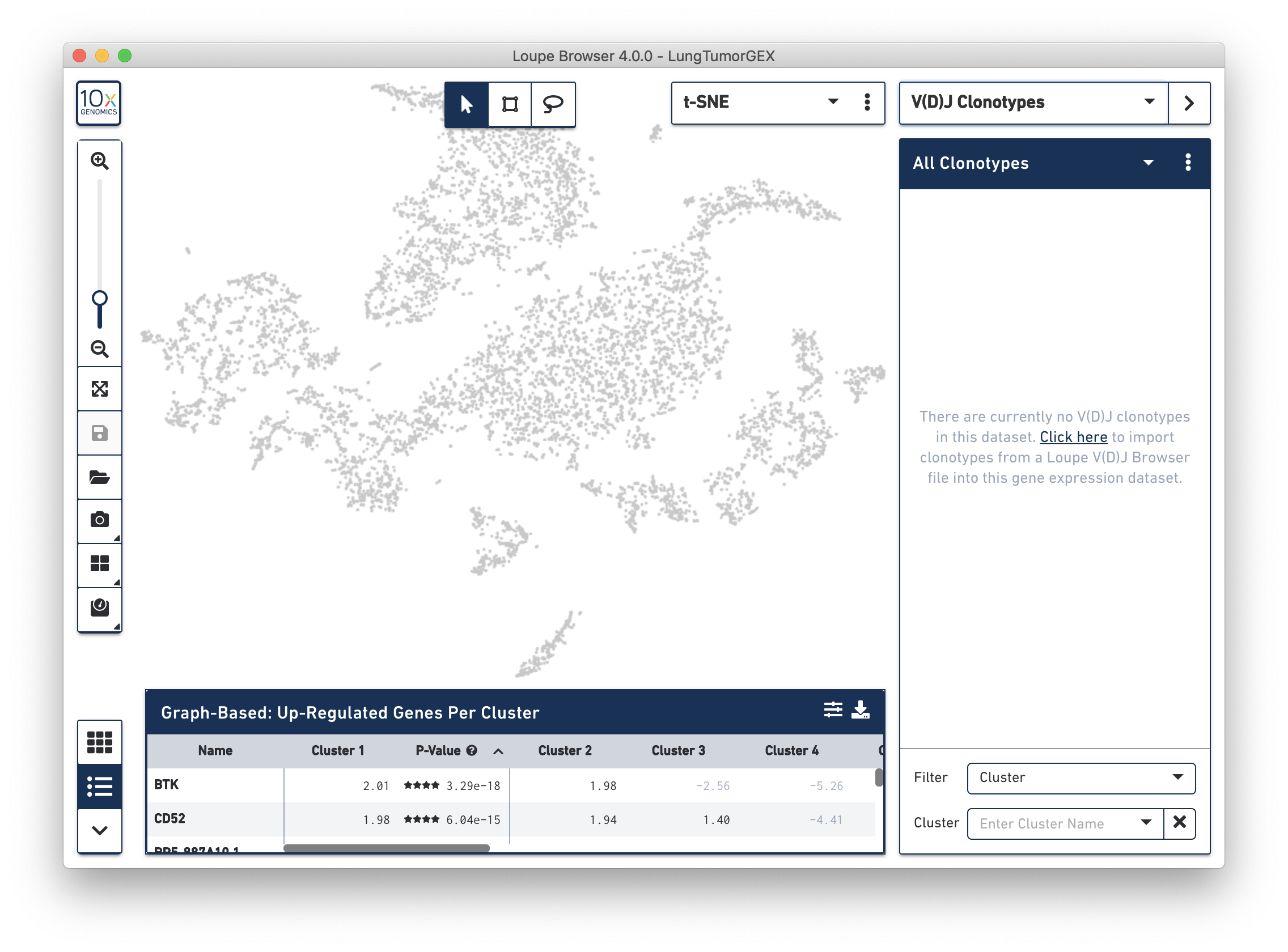
In V(D)J Clonotypes mode, you can load an input sample's corresponding V(D)J data into the Loupe Browser workspace. Cells that belong to the active set of V(D)J clonotypes are colored blue, and the sidebar changes to a filterable clonotype list. For more information on how to use Loupe Browser to combine gene expression and immune repertoire data, follow the Integrated Gene Expression and V(D)J Analysis in Loupe Browser tutorial.
The data panel on the bottom of the workspace displays information about the features driving differences between clusters. By default, it enumerates the genes that drive differences between the current precomputed clustering selected in the Categories sidebar. It also displays the results of a Significant Features analysis. The mini-toolbar to the left of the data panel switches between a table and a hierarchical heat map of differentiating features. Clicking on a feature name in the table view allows you to add that feature to a list for future reference, or to show counts of that feature across the dataset in the active projection.