Cell Ranger4.0, printed on 04/04/2025
This tutorial reviews the major analysis capabilities Loupe Browser provides for analyzing the following data:
To use Loupe Browser, follow the directions on the Installation page to download and install the software on either macOS or Windows.
Navigating the Loupe Browser User Interface
If you are new to Loupe Browser, learn more about the capabilities in the Navigation Tutorial.
Once you have installed Loupe Browser and are familiar with the basic navigation capabilities, you can start the tutorial to learn how to analyze data. Open Loupe Browser by double clicking on the application icon. Then click on the AMLTutorial.cloupe file from the list of Recent Files.
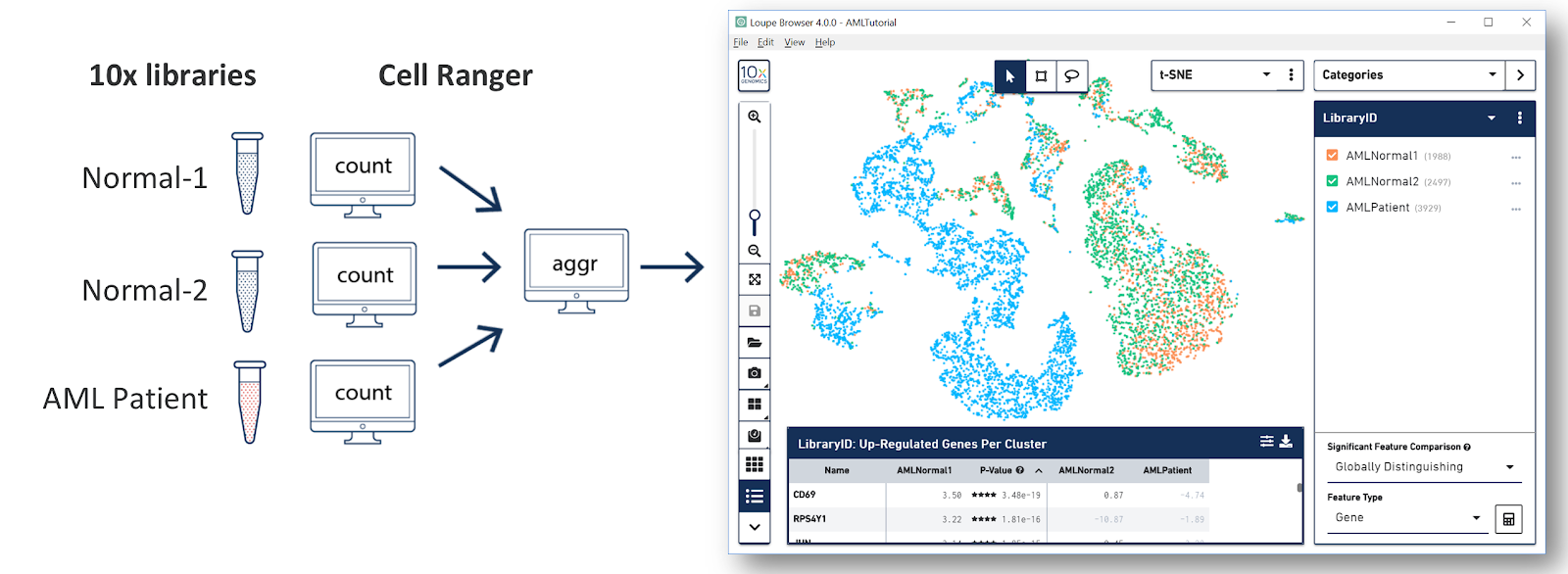
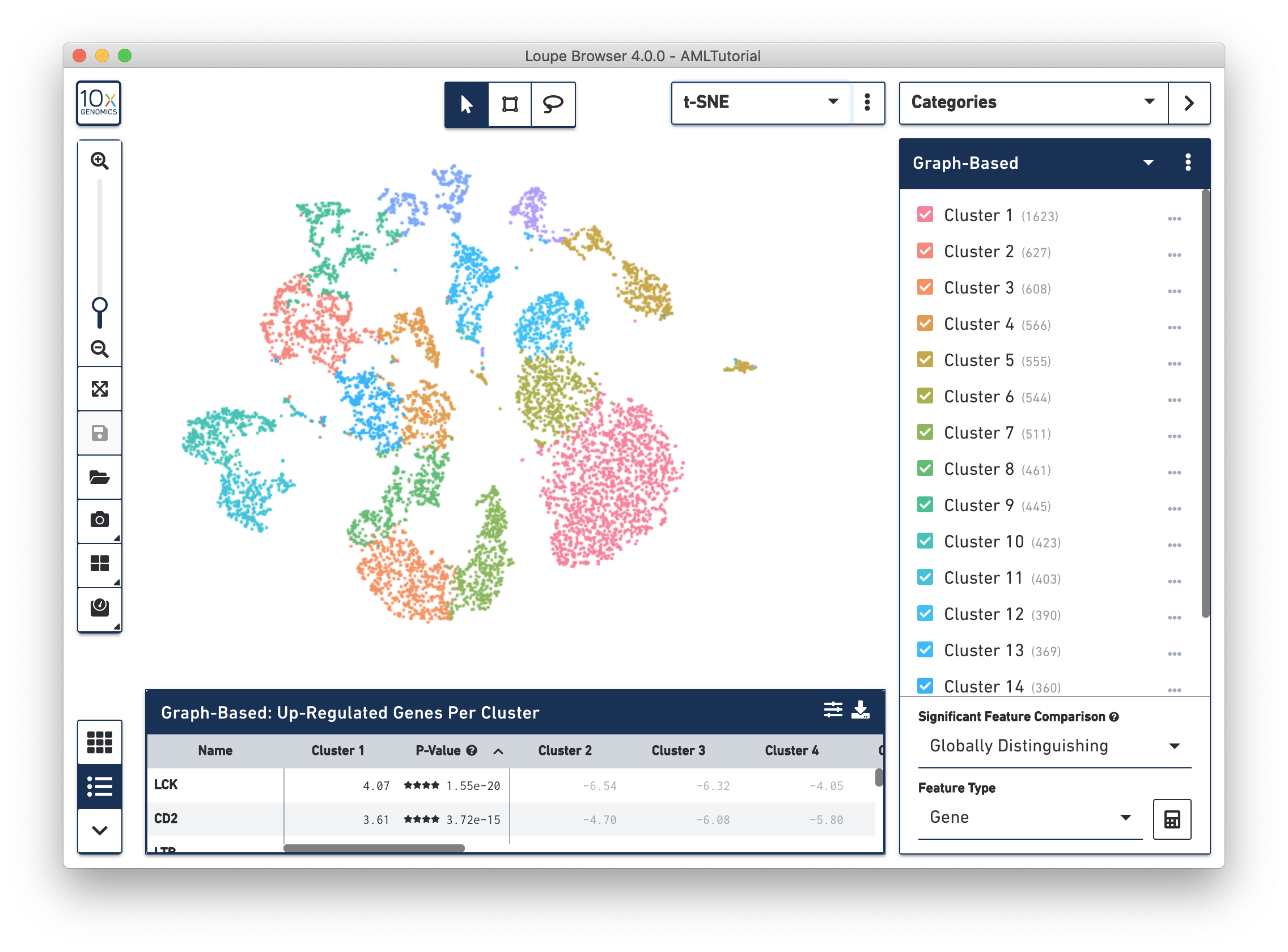
Once you do that, you see a screen with the t-SNE projection for the cells from the AML dataset.
The AMLTutorial dataset contains the results of a Cell Ranger aggr run over three samples, including two healthy control samples of frozen human bone marrow mononuclear cells, and a pre-transplant sample from a patient with acute myeloid leukemia (AML). This dataset was generated in collaboration with the Fred Hutchinson Cancer Research Center, and referenced in the Nature Communications publication, "Zheng et al, Massively parallel digital transcriptional profiling of single cells" (2017; doi:10.1038/ncomms14049).