Cell Ranger5.1, printed on 04/01/2025
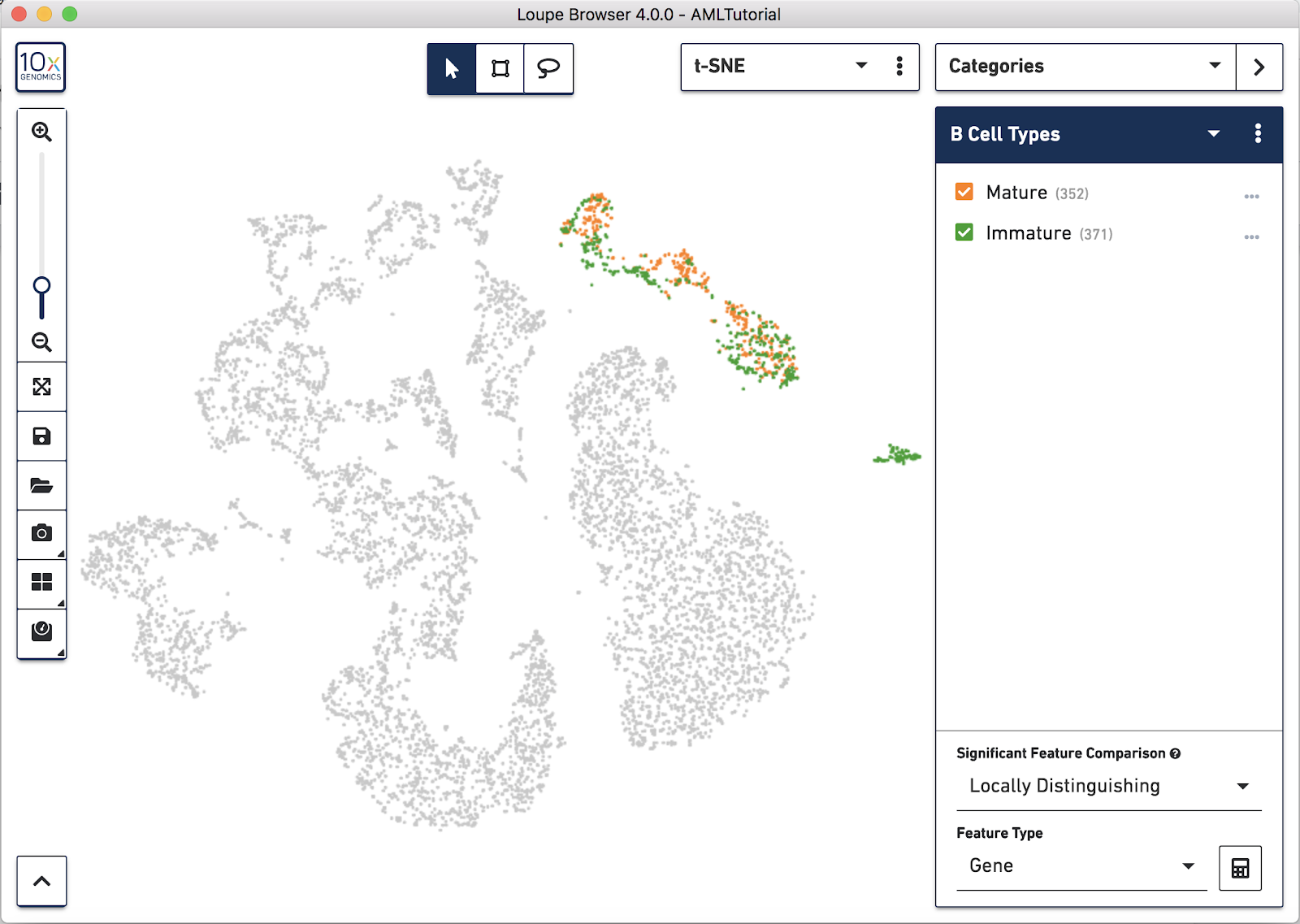
Goal: To identify known immature and mature B Cell types in the dataset.
Let's build on the work we've done in the identifying cell types and exploring substructure sections. We'll use the Filters panel available in Loupe Cell Browser 3.1 or later or versions of Loupe Browser 4.0 and higher.
In the identifying cell types section we identified B Cells with marker genes and in the exploring substructure section we further segmented B Cells in t-SNE space by lassoing. Let's find and create new clusters of immature and mature B Cells within the previous B Cell cluster we found by using the presence or absence of TCL1A.
Let's open the Filters interface in the right panel and start building a filter. First, we'll add all three clusters that we created in exploring substructure. You can add a cluster by clicking the "Create New Rule" button, then the "in [cluster name]", and finally enter the cluster name "All Cells". Repeat this with the two other clusters: "Cluster 1" and "Cluster 2". Since these two cluster names are shared among multiple categories, you can prepend the category name "B Cells" to the cluster name, or find the cluster in the dropdown that belongs to the "B Cells" category which appears under the cluster name.
You may have noticed that after adding all three clusters, there are no barcodes selected. You may have also noticed that there is a human-readable form of the filter at the bottom of the panel which updates whenever a change is made. It should currently show that the ruleset is filtering for barcodes in All Cells and Cluster 1 and Cluster 2. However, there exist no barcodes that are in all three (or even two) clusters. Instead, we want to find barcodes that are in either All Cells or Cluster 1 or Cluster 2. We can accomplish this by clicking the toggle in the top right of this ruleset. You should immediately see that the clusters in the top right are now highlighted in purple, the same clusters that we created in exploring substructure.
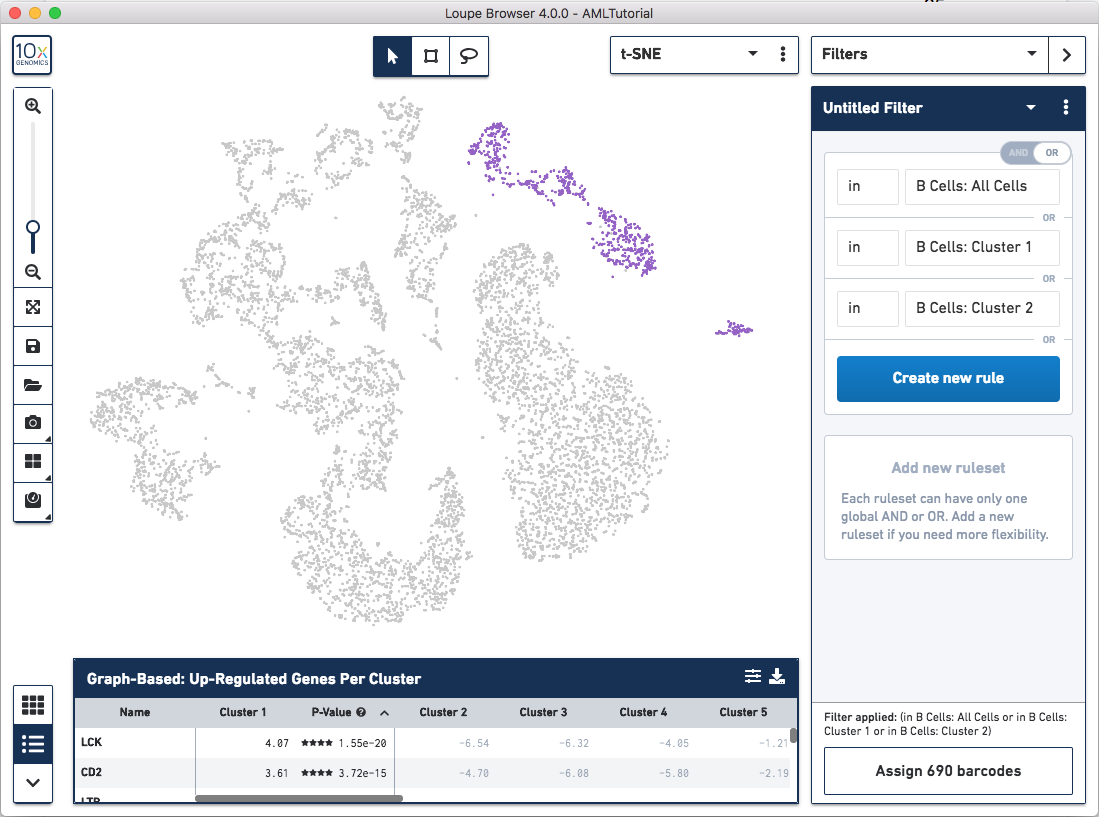
Next, to find immature B Cells, we want to find barcodes that are in these
clusters and expressing TCL1A. Since each ruleset is restricted to a single
boolean operator, we need to add a new ruleset. Click the "Add new ruleset"
button and then click the new "Create new rule" button. Add a rule that
thresholds on count that's greater than a value by clicking the > button in the
"Threshold by count" section. For this rule, select the gene TCL1A. You should
see a subset of the barcodes that were highlighted which should represent
immature B Cells. The human-readable filter at the bottom should now read:
(in B Cells: All Cells or in B Cells: Cluster 1 or in B Cells: Cluster 2) and (TCL1A > 0).
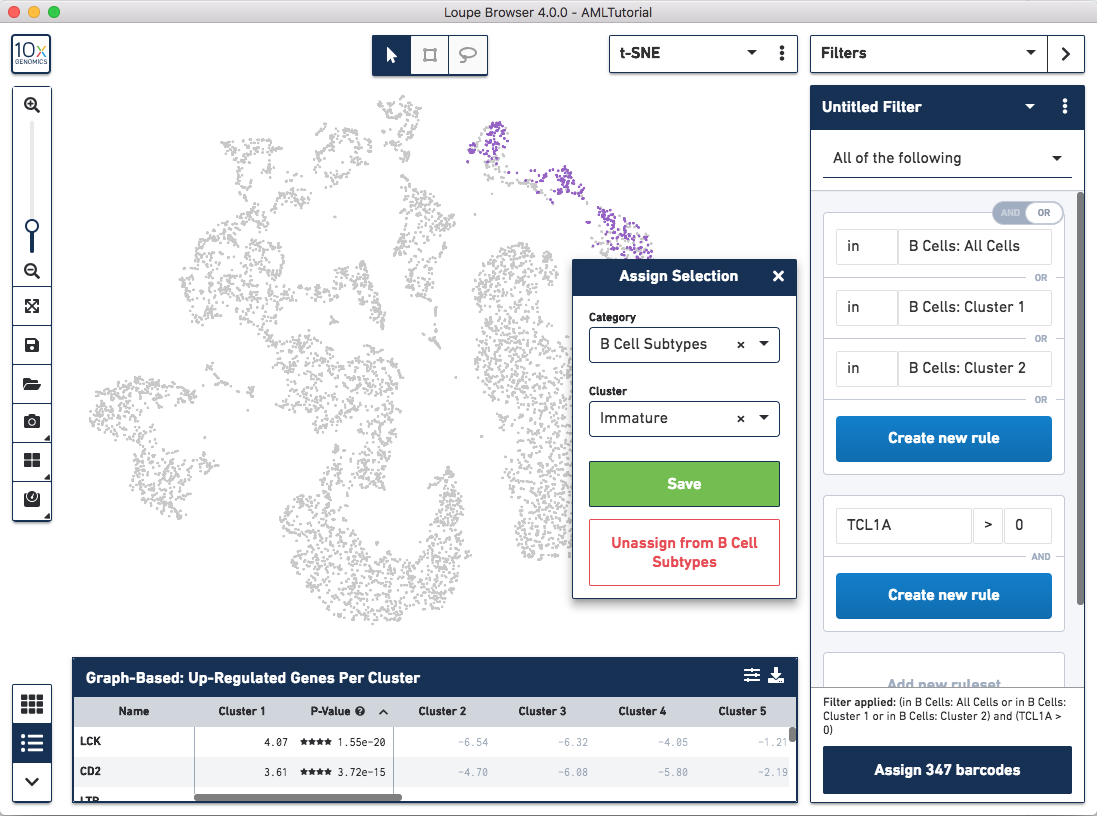
Let's assign these barcodes to a new category by clicking the "Assign 347 barcodes" button at the bottom. Name the category "B Cell Subtypes" and the cluster "Immature".
Now, let's find mature B Cells by modifying the filter we just made. Since mature B Cells don't express TCL1A, let's modify the existing TCL1A rule we created so that the rule reads "TCL1A = 0" instead of "TCL1A > 0". You can do so by clicking the greater than symbol (>) directly and choosing equals (=) instead. You should see a different highlighted subset of B cells in purple. Assign these 343 barcodes to the same category "B Cell Subtypes" and name the cluster "Mature".
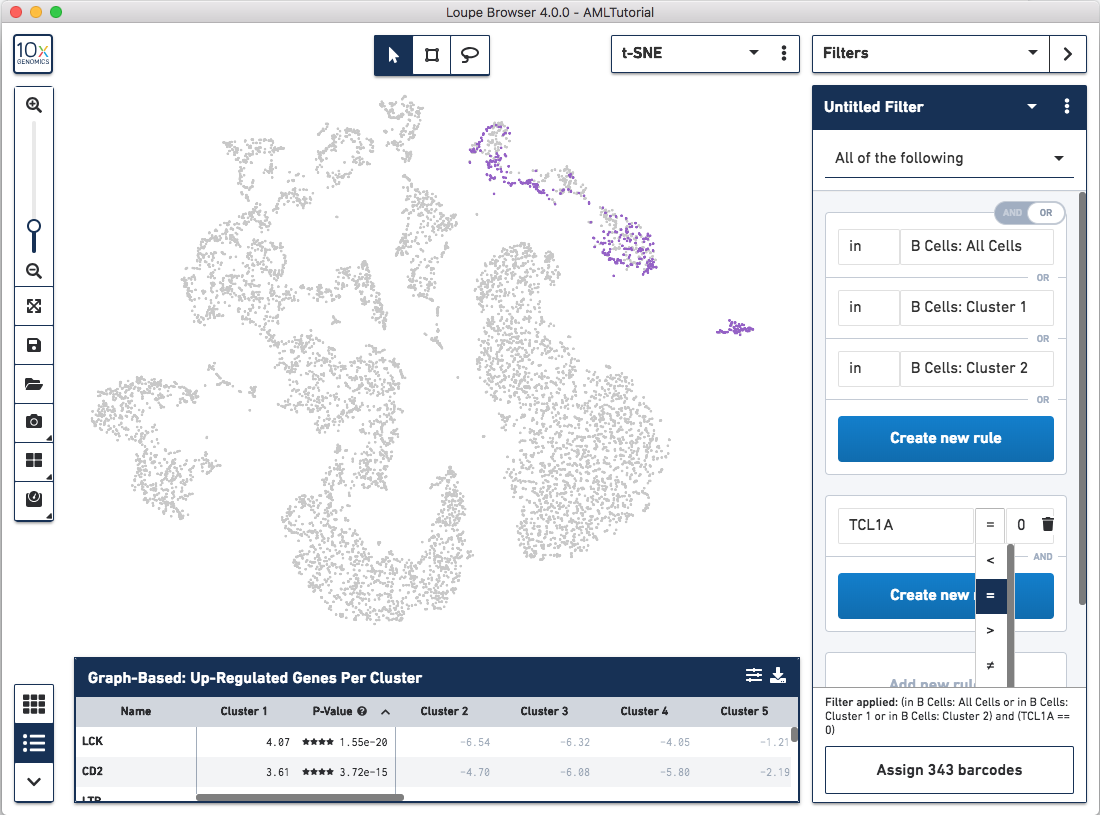
Now that you've segmented the B cells in this dataset, you can choose to run differential expression between these subtypes like we did in exploring substructure. It's interesting to note that "Cluster 2" (the bottom right-most cluster) of the B Cells categories looks like it's exclusively mature B Cells, while the other two are mixed.
Filters allow you to create boolean filters with one level of nesting. Each ruleset represents a grouping of rules which are joined by the same logical operator. The logical operator can be toggled between AND/OR using the slider in the top right of the ruleset.
You can add additional rulesets when the first ruleset has more than one rule by clicking the "Add new ruleset" at the bottom. Rulesets operate independently from one another. Each ruleset may have its own logical operator.
Rulesets are all joined by the same logical operator. This can similarly be toggled between AND/OR using the dropdown at the top when there are greater than two rulesets. The options are:
In the human-readable form of the filter at the bottom, each ruleset is grouped with parentheses. The logical operator within parentheses must be the same operator. Similarly, the logical operator between sets of parentheses (rulesets) must be the same operator.
We've now done some substantive exploration of this dataset. Let's finish the tutorial by learning how to save and share our work.