Space Ranger6.4, printed on 03/31/2025
Space Ranger 2.0 introduced support for CytAssist-enabled Gene Expression analysis which uses the instrument-generated image on a CytAssist slide along with optional microscope image of the same tissue section on a standard glass slide. The pipeline uses automatic image registration algorithm to align and optimize the overlay of the two image inputs. While the automatic image processing pipeline is very robust, in cases where there are shape mismatches (e.g. missing portions of the tissue section, excessive folding or tearing), or stain mismatches (e.g. excessive stain leakage or incomplete destaining or weak re-staining) in the tissue section between the CytAssist and microscope image, manual image registration will be required.
Loupe Browser 6.2 onwards provides the CytAssist Image Alignment wizard to achieve manual image registration. The alignment wizard makes it possible to interactively upload the two image inputs, select key landmark points between the images, and algorithmically refine the alignment. The alignment wizard provides an option to export a standalone alignment file with image registration results, or to continue with the manual fidual alignment workflow to generate a combined alignment file. This tutorial walks through the image registration workflow step-by-step.
The Visium manual aligment wizard can be found on the Loupe Browser start page, at the bottom of the main panel.
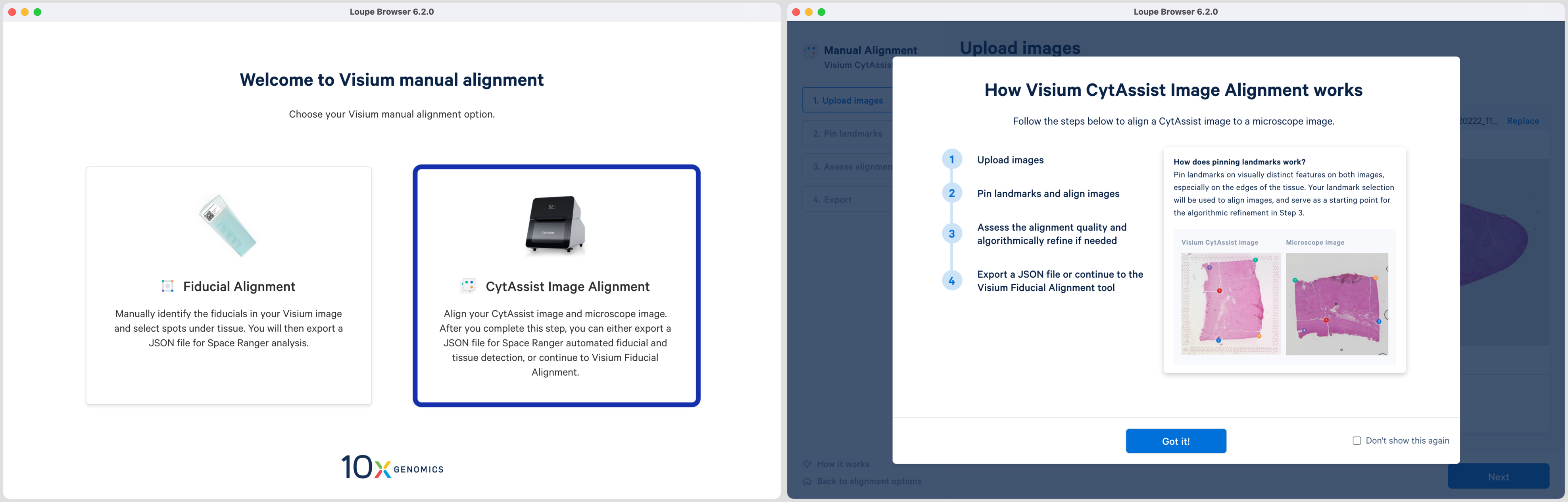
Clicking on the link starts the alignment wizard in a new window and shows a pop-up window listing the two available workflows. Click CytAssist Image Alignment box to open a pop-up window which summarizes the steps required for completion. Click to exit out of the pop-up window. Selecting the box for Don't show this again will bypass the pop window the next time this alignment wizard is started.
| The images uploaded in the CytAssist image alignment wizard must match the images passed to the spaceranger count pipeline for analysis. |
There are two main image inputs: the CytAssist instrument generated image and the microscope image (brightfield or fluorescence), which follow the same format requirements as that for Space Ranger. The CytAssist image, which contains the fiducial frame, has fixed resolution and size specifications which are used by both Space Ranger and Loupe Browser to verify the image type. Loupe 6.2 also adds support for .qptiff image format. . Refer to the input image recommendations page for more details.
| Image Format | |
|---|---|
| CytAssist Image | 24-bit color TIFF |
| Microscope Brightfield Image | 24-bit color TIFF/BigTIFF or a JPEG |
| 16-bit grayscale TIFF/BigTIFF or a JPEG | |
| Microscope Fluorescence Image | 8 or 16-bit grayscale single, multi-channel TIFF/BigTIFF |
| 8 or 16-bit grayscale single-channel TIFF/BigTIFF or a JPEG | |
| 24-bit single colored image TIFF/BigTIFF or a JPEG |
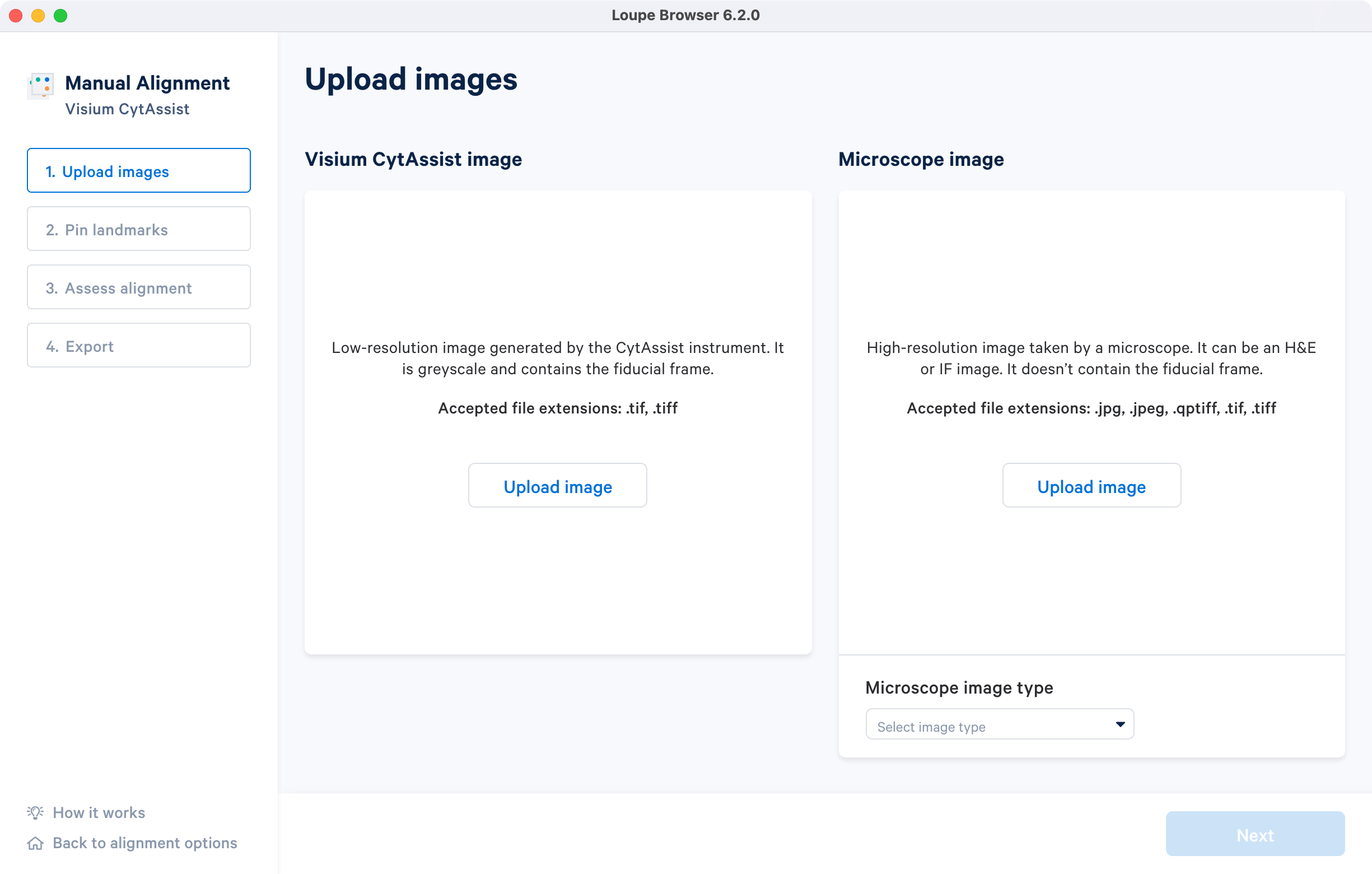
Clicking on under the Image for alignment panel opens a file dialog that filters the current filesystem for compatible image files. Selecting a file triggers a brief loading process. Note that clicking on the How it works on the lower left corner brings up the pop-up window.
Select the tab corresponding to the different starting image types.
| To minimize download time, the example images included in this tutorial are downsampled versions of the images included in the CytAssist public datasets containing Brightfield and Fluorescence image pairs. |
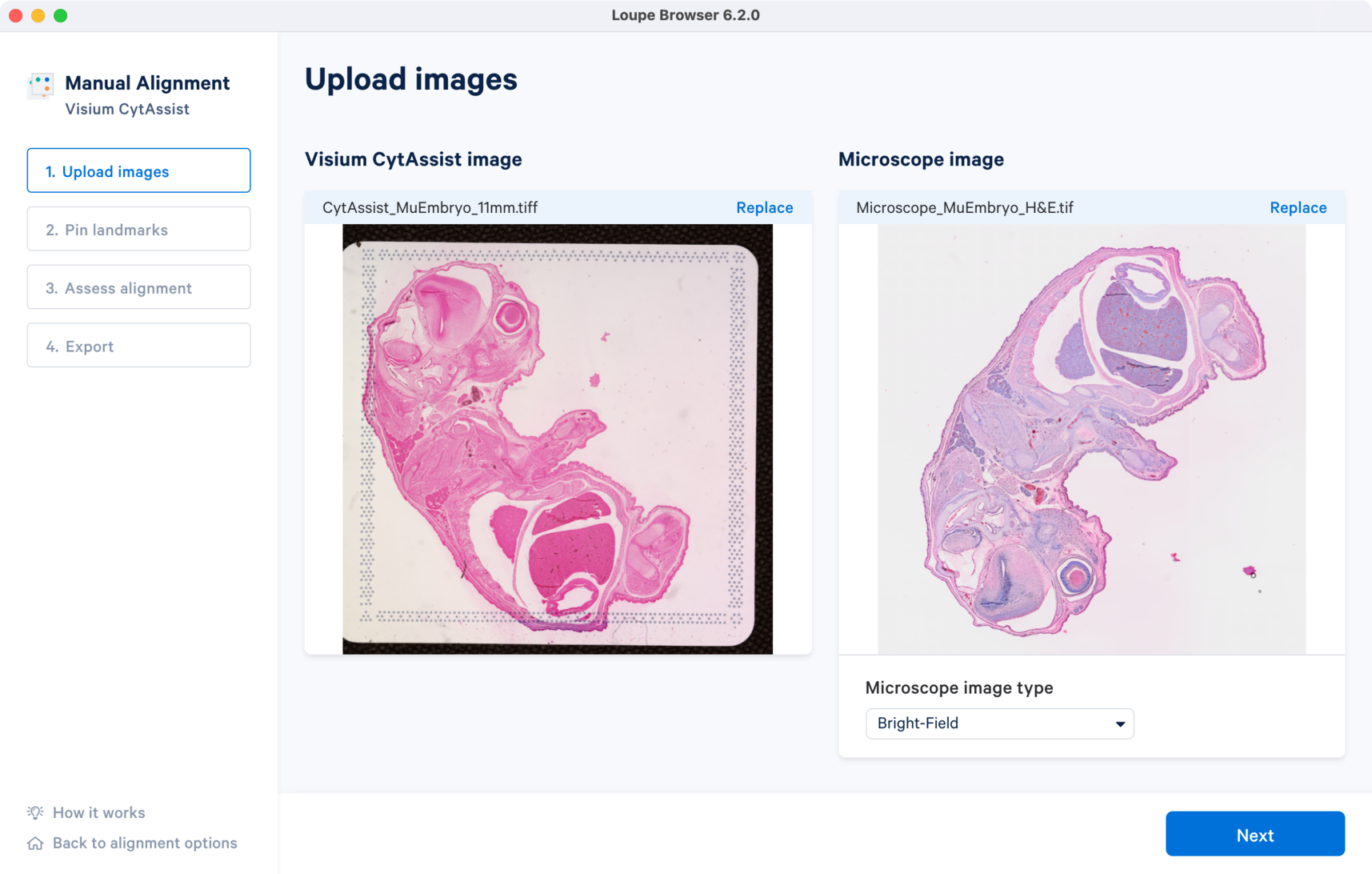
The tutorial will use the following resources for the CytAssist image and H&E-stained brightfield microscope image.
Once the image is loaded, preview image appear, as shown below. After successful upload of the images, select the image type from the Microscope image type drop down menu which will make available for click. Use the Replace option located at the top right corner above image preview to change the loaded image.
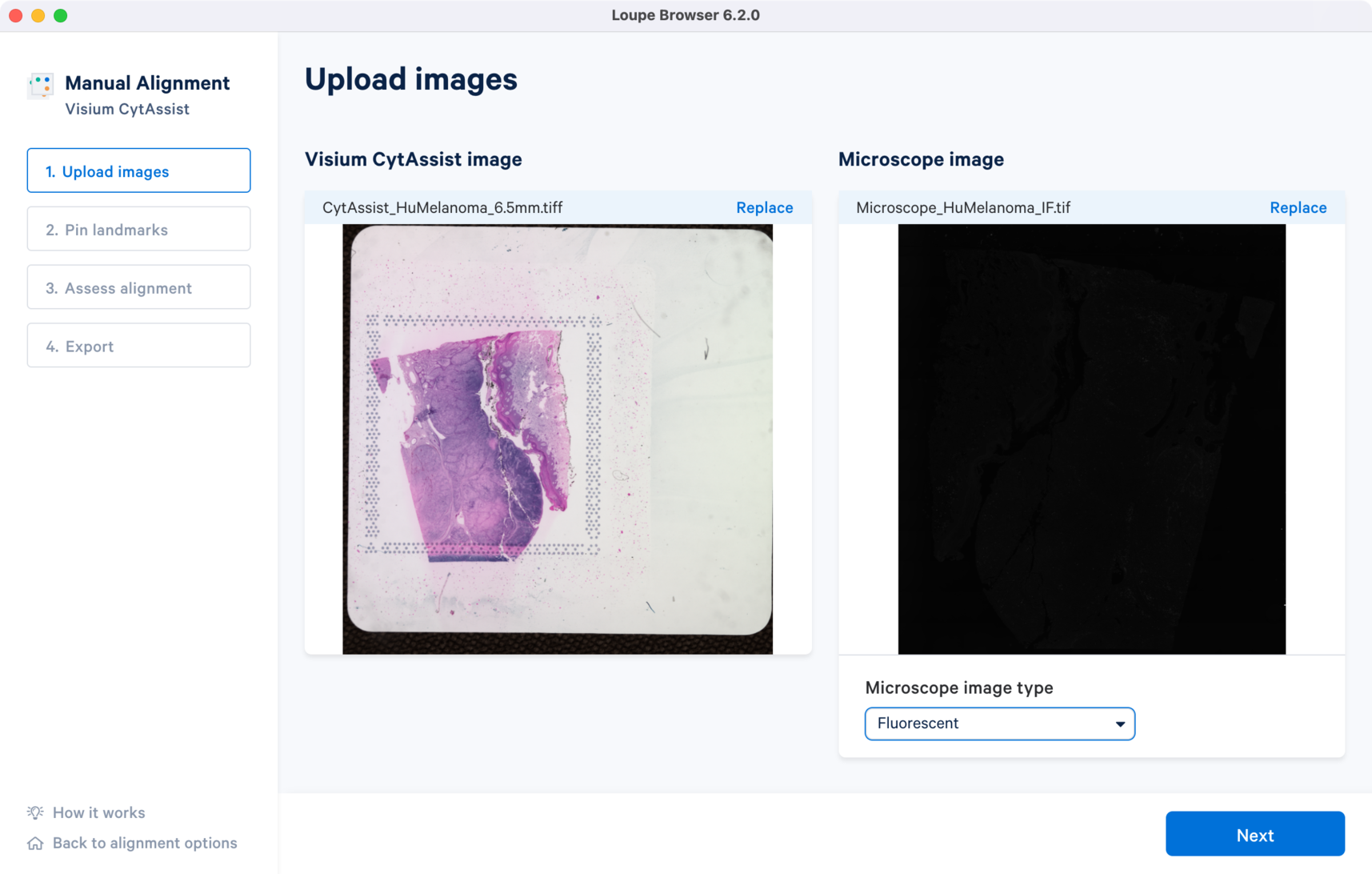
The tutorial will use the following resources for the CytAssist image and multi-channel immunofluorescence microscope image.
Once the image is loaded, the preview image appears, as shown below. After successfully uploading the images, select the image type from the Microscope image type drop down menu which will make available for click. Use the Replace option located at the top right corner above the image preview to change the loaded image.
For multi-channel immunofluorescence image, the Image channel dropdown to change the active channel selection will be available in Step 2. Pin landmarks.
The success of the manual image registration is dependent of accurate landmark selection between the two images. Since the initial overlay of the two images is derived from the landmark placements, there are some key considerations for choosing landmarks:
Click in the alignment wizard to access a summary of these key points along with example images.
Select the tab corresponding to the different starting image types. Click to undo the landmark selections.
Since the resolution between CytAssist and microscope image will be different with CytAssist representing a lower resolution version of the tissue section, the zoom applied to each image can be independent when selecting landmarks.
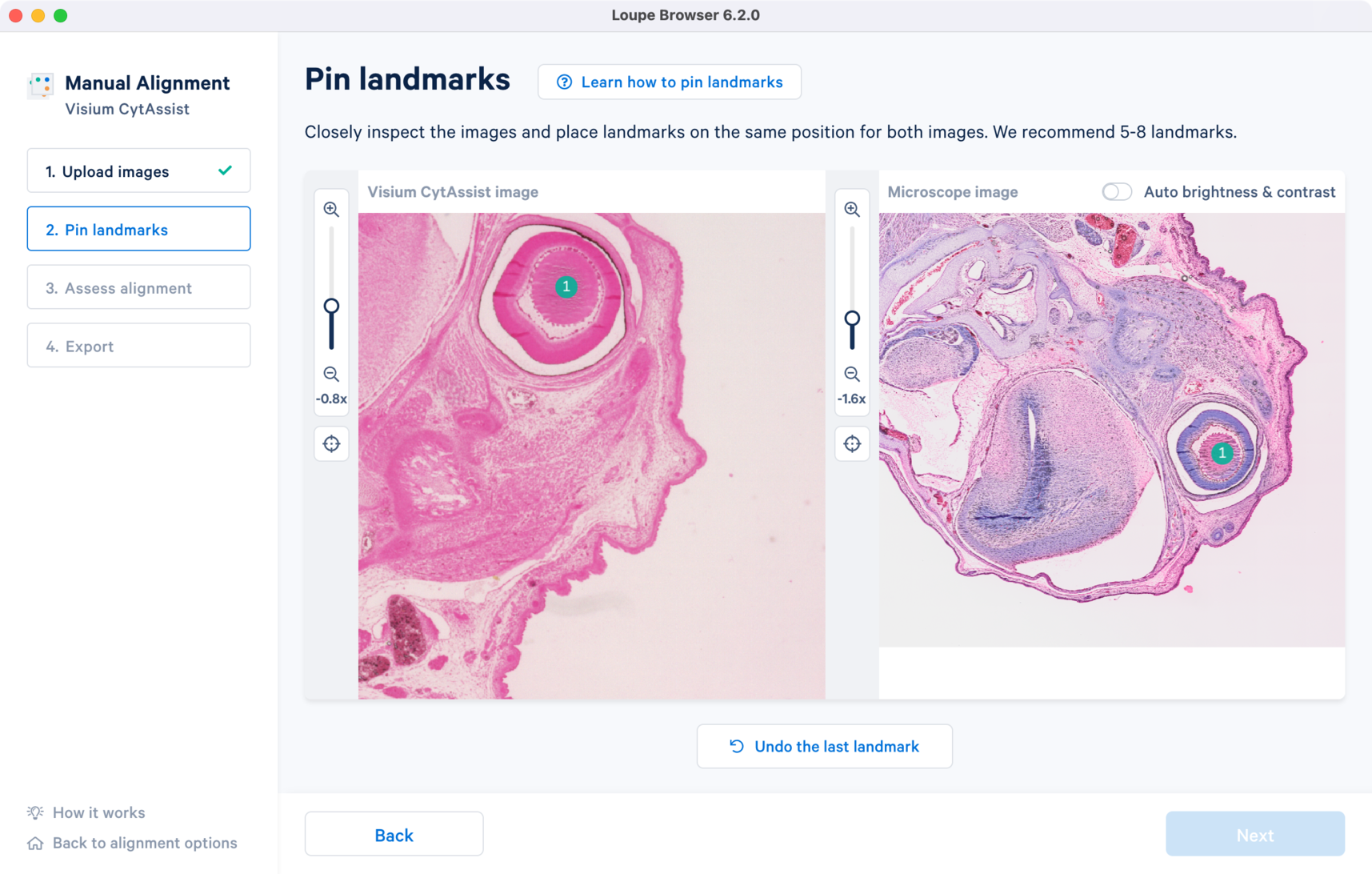
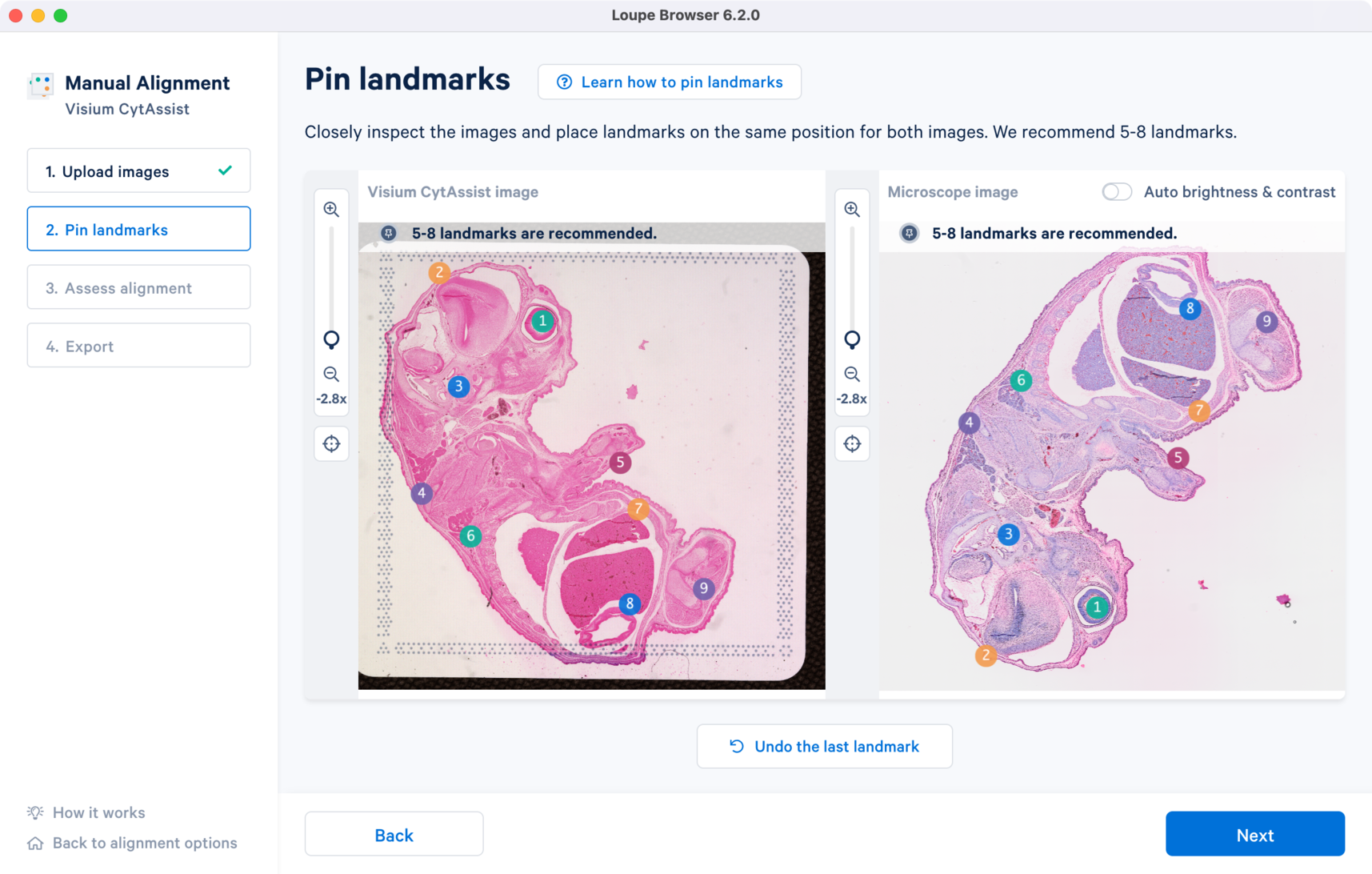
The landmarks are selected based on the above criteria. Click to zoom out and view the final landmark placements across both the images. Click to proceed to Step 3 Assess alignment to view the image registration results.
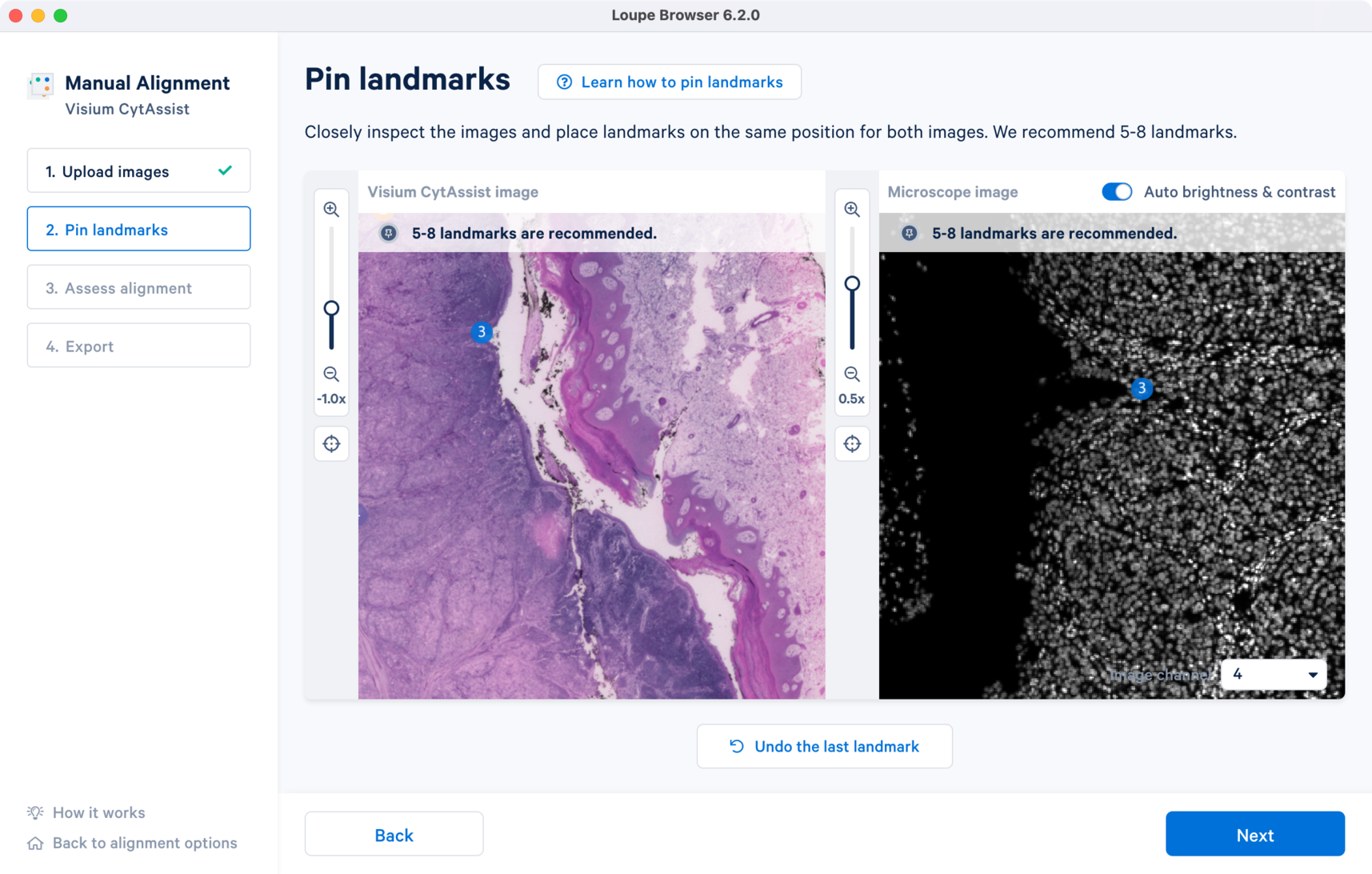
For enhanced visualization, turn on the Auto brightness & contrast which applies uniform brightness scaling across all pixels in the image. This is a useful feature for dark background fluorescence images.
For multi-channel fluorescence images, a drop-down menu for the Image channel selection is available for the microscope image. All landmarks chosen in one channel carry over when the channel view is switched. This is particularly useful because the morphological features captured by each of the antibody stain may be distinct.
Since the resolution between CytAssist and microscope image will be different with CytAssist representing a lower resolution version of the tissue section, the zoom applied to each image can be independent when selecting landmarks.
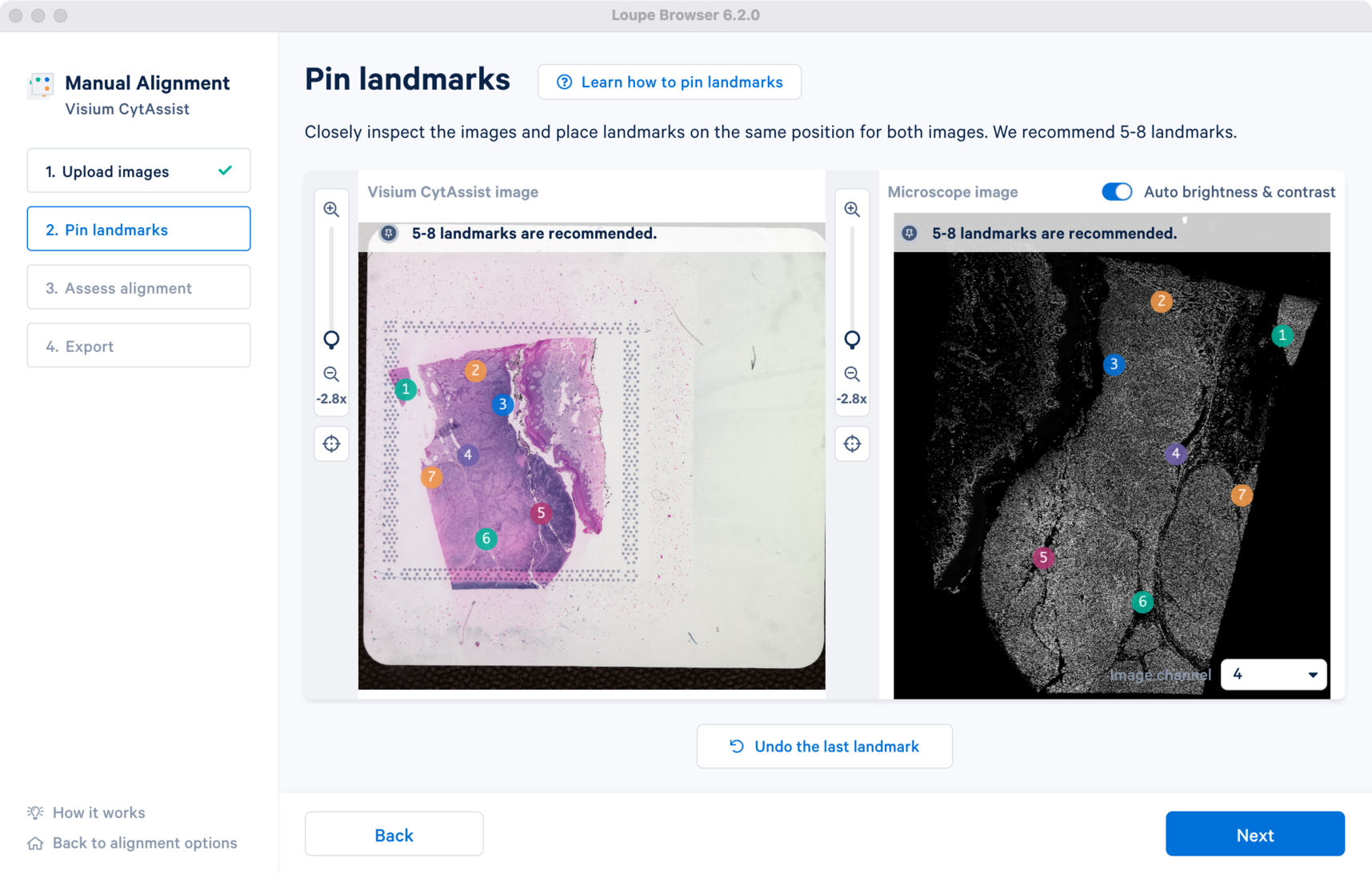
Click to zoom out and view the final landmark placements across both the images. Note that the channel view set at this step will be used in the Step 3 Assess alignment
This step provides the results of the image registration based on the landmark selection from Step 2. Pin landmarks.
In the window are the overlap of the two image inputs. The first alignment is based only on the landmark selection. An opacity slider is provided on the right hand side which is set to 50% value as default. To assess the quality of the registration, zoom into the image and use the opacity slider to scroll in either directions to examine the overlap between the edges as well as unique tissue features between the two images. When there are no shape discontinuity or shift of one image relative to another, the images are considered to be successfully reegistered.
Click in the alignment wizard to view some example images of good and bad alignment cases.
Continuing with the tutorial examples, for the set using CytAssist and Fluorescence microscope images, the initial inspection shows good alignment between the two images. In this case, the results are satisfactory and can be exported for analysis in Step 4. Export.
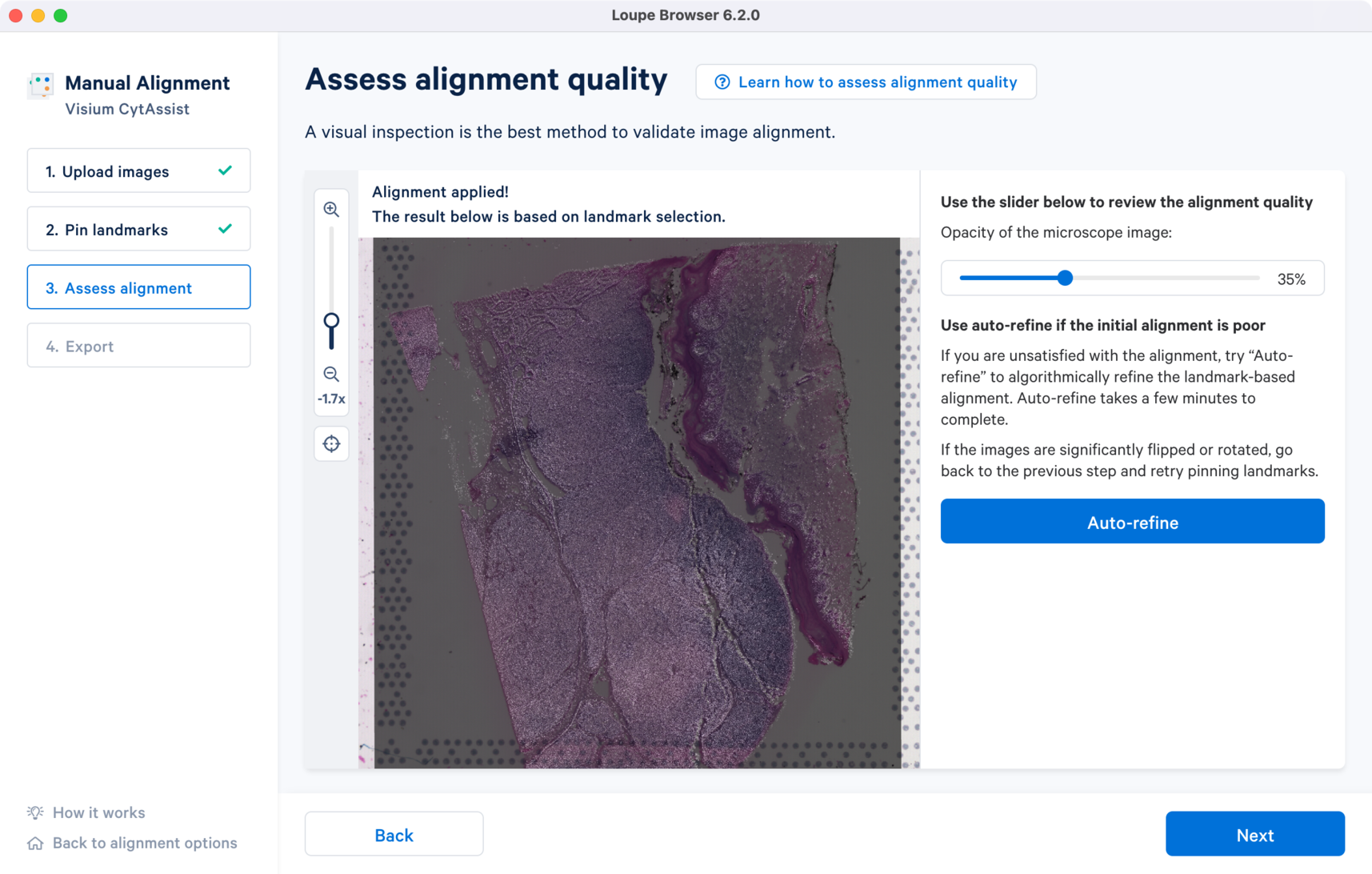
For the CytAssist + Brightfield image pair, the initial alignment looks close with some room for improvement around the edges. There are two ways to refine the alignment:
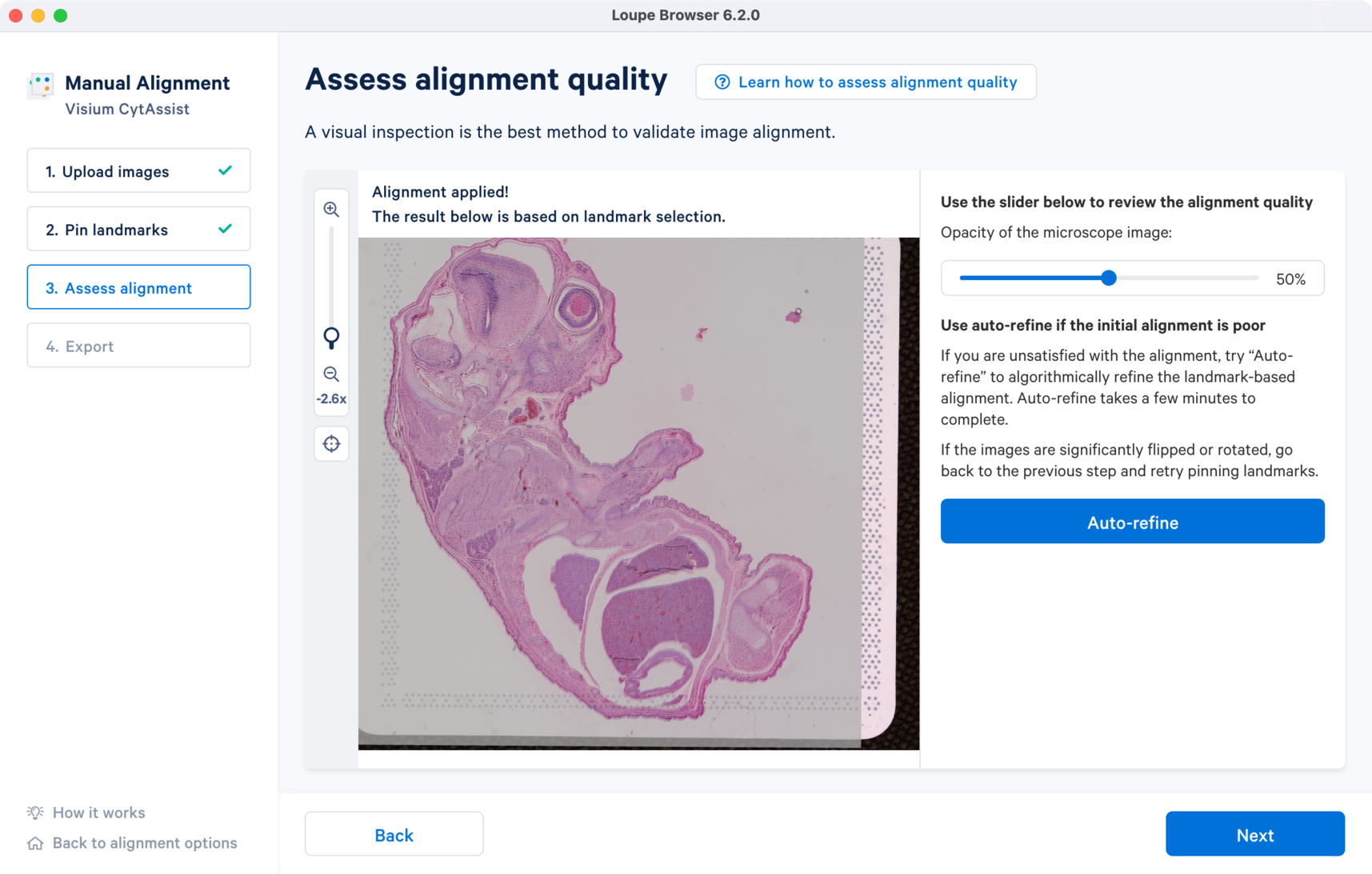
In this tutorial, the first option is choosen. Auto refinement algorithm optimizes the image registration based on the landmark-based initial state. Since it is an iterative process, it can be applied multiple times on the same image pair.
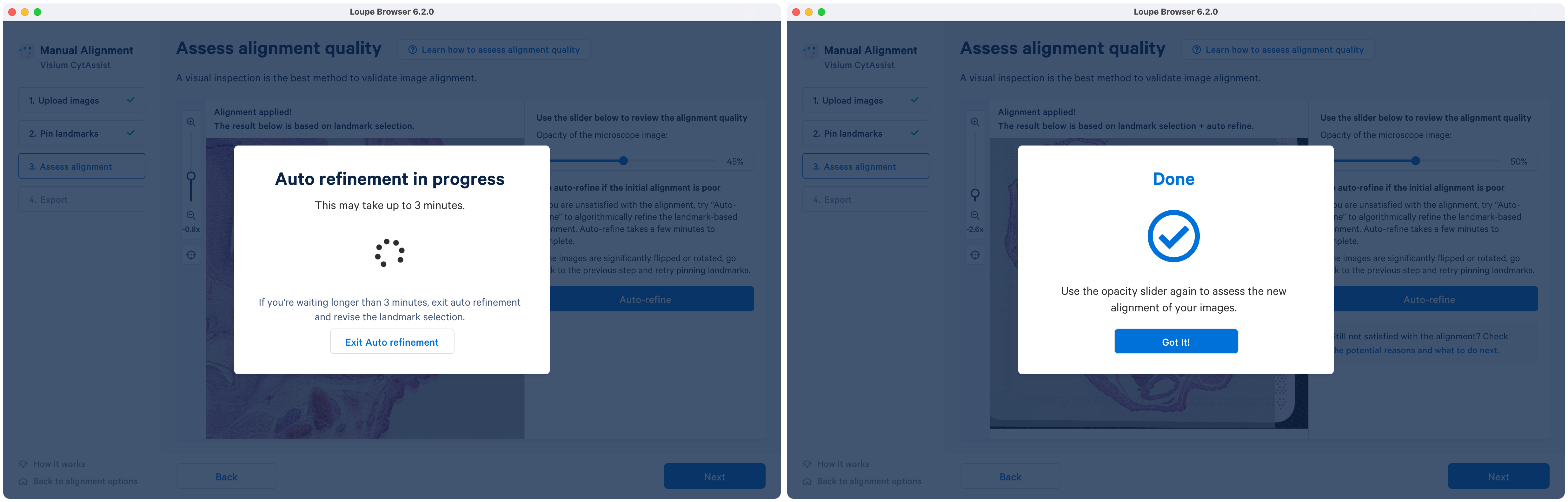
Clicking opens a new pop-up that shows the active state of the refinement with a spinning circle. Upon completion the text on the window is updated and to exit click .
Although there is an option to exit out of the auto refinement pop-up window, it is highly recommended to allow the algorithm to finish. In cases where the algorithm runs longer than indicated, it suggests a potential problem with the initial alignment state. In those cases, it is advisable to exit, and redo the landmark selection for more accurate image registration. Potential reasons and guidance for next steps can be found by clicking on the potential reasons and what to do next in the alignment wizard.
The image registration is significantly improved after applying the algorithm. Notice the overlap distance between the two images along the edges has reduced from the initial landmark based alignment. The results are satisfactory and can be exported for analysis in Step 4. Export.
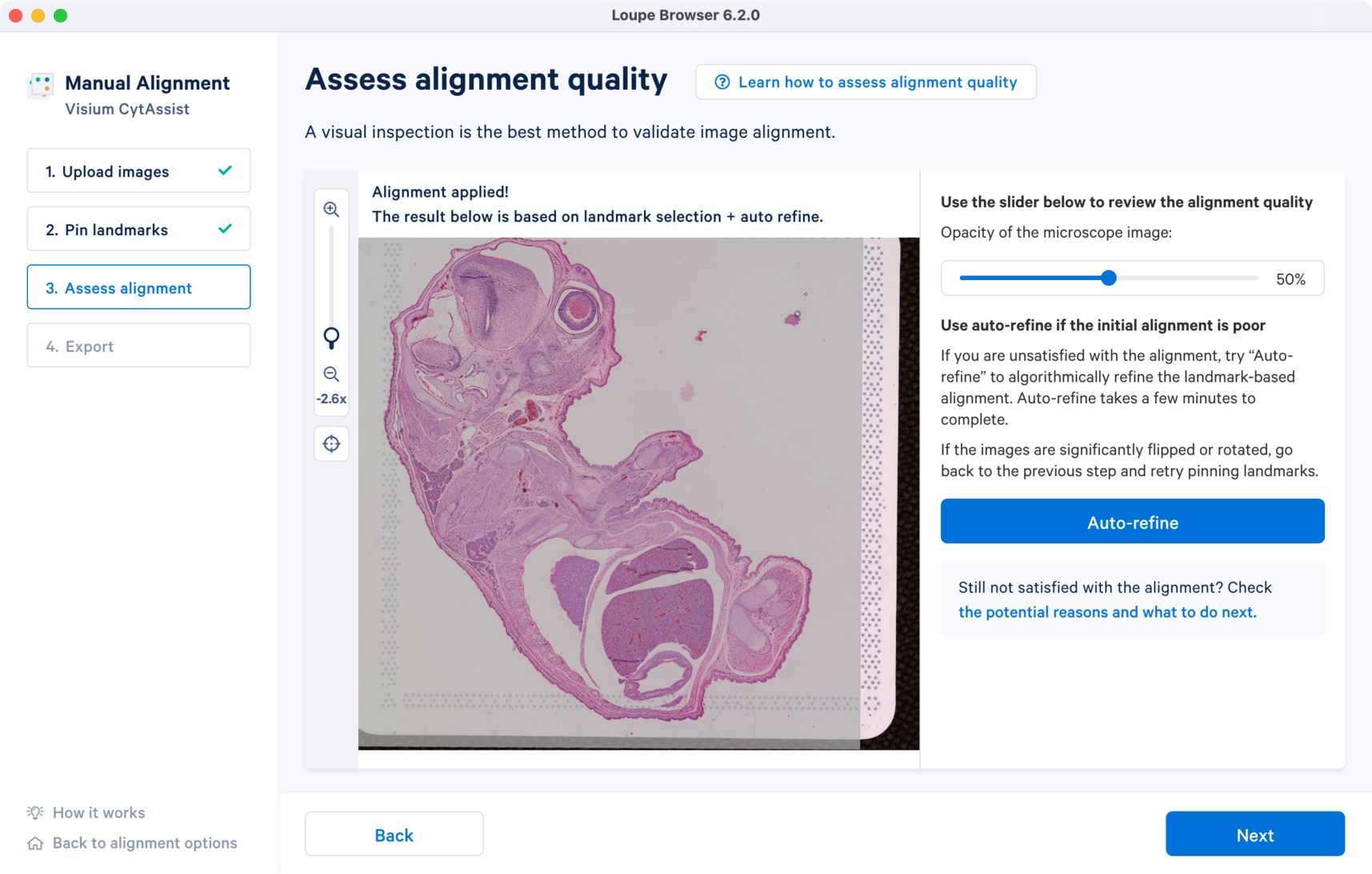
The last step in the CytAssist Image Alignment wizard provides two options: 1) export the image registration results in a JSON file format or 2) continue to the manual fiducial alignment wizard to manually align the fiducial frame and segment the tissue section and generate a combined JSON file. More details about the different manual workflow options are listed in the spaceranger count for CytAssist analysis page.
| Since the CytAssist image contains the fiducial frame, it is always used for the manual fiducial alignment workflow irrespective of the microscope image format (brightfield or fluorescence). The brightfield CytAssist image is compatible with the automatic fiducial detection and tissue segmentation pipeline in Space Ranger. |
The CytAssist images used in this tutorial have visible fiducial frame that is largely unobstructed, and the strength of the tissue staining is sufficient to identify morphological features. In these cases, the alignment file can be exported by clicking the to generate a JSON file.
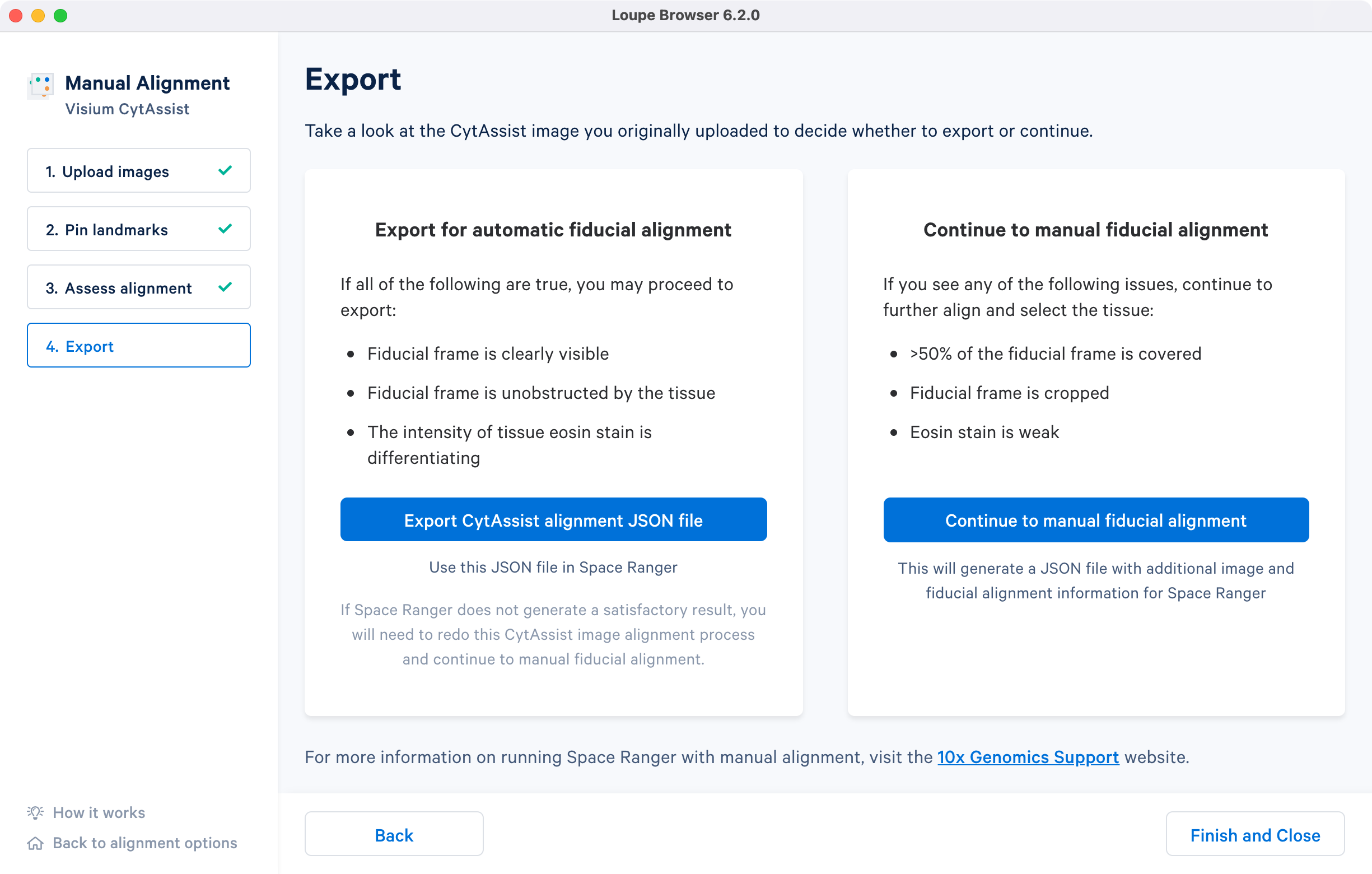
If the CytAssist image has partial fiducial frame obstruction by the tissue section or one of more edges were cropped, then it warrants continuing with the manual fiducial alignment workflow. Some subset of issues with the tissue staining (weak staining, incomplete staining, or excessive staining with leakage outside of the tissue section) can also interfere with accurate identification of the tissue associated spots using the automated image processing pipeline. In these cases, it is recommended to complete the manual fiducial alignment workflow.
Click which will automatically load the CytAssist image from the previous step. Follow the instructions in the manual fiducial alignment tutorial to complete the steps in the alignment wizard. The alignment file generated at the end now contains information about both the image registration as well as the fiducial detection and tissue segmentation.
The path to the generated JSON file is provided to spaceranger count pipeline via the --loupe-alignment argument. For more information, consult the Manual Alignment instructions in the Space Ranger documentation.
Space Ranger does not support specifying multiple alignment files. If the pipeline is run using the alignment file containing only the image regstration information and the results are not satisfactory, both the manual workflows must be repeated i.e the CytAssist Image Alignment and the Fiducial Alignment workflows. The combined JSON is then used to re-run the pipeline. |