Space Ranger2.0, printed on 03/30/2025
| This page lists the resources for analysis of whole transcriptome profiling using Visium Spatial Gene Expression for FFPE direct placement workflow in formalin-fixed paraffin-embedded (FFPE) samples. Refer to the corresponding pages for direct placement workflow for Fresh Frozen (FF) samples using poly-A based assay and CytAssist enabled workflow for either FFPE or FF samples using probe-based assay. |
The Visium Spatial Gene Expression for FFPE workflow measures RNA levels in intact, formalin-fixed paraffin-embedded (FFPE) tissue sections, using probes targeting the whole transcriptome. After whole transcriptome probe panels are added to the tissue, each probe pair hybridizes to its target gene and is then ligated. The ligation products are released from the tissue and bind with spatially barcoded oligonucleotides present on the capture area. Libraries are generated from the barcoded probes and are sequenced. The final sequencing library structure is similar to Visium for fresh frozen tissue, but the synthetic probe DNA is sequenced rather than cDNA of a captured transcript. For FFPE samples, Space Ranger counts oligo ligation events to build the feature-barcode matrix. As a consequence, if the probe hybridizes to a transcript with a variant, that variant will not be present in the sequencing data. The probe design avoids known SNPs.
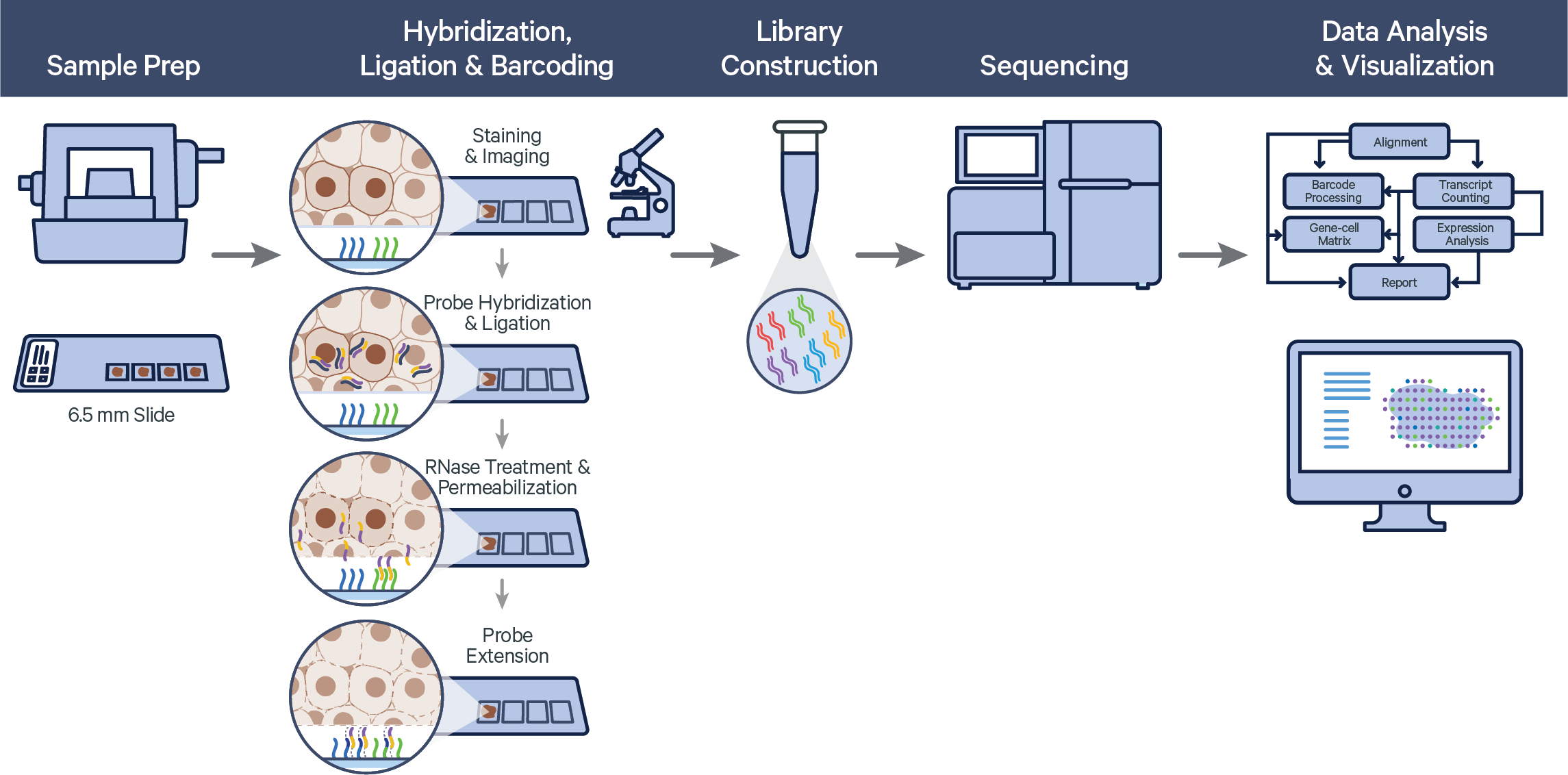
Visium Spatial Gene Expression for FFPE relies on the same underlying Space Ranger analysis software as Visium Spatial Gene Expression, but uses a short read aligner tailored to probe sequences. The probe aligner assigns probe and gene IDs to properly aligned reads, and differentiates reads originating from ligation of correctly paired probe halves from potential artifacts and non-probe reads.
Resources and additional information are available below: