Space Ranger2.0, printed on 03/30/2025
Image input types and formats at a glance. Considerations for each of these image types are detailed below.
| Image Format | Space Ranger Flags | Automatic Fiducial Alignment | Automatic Image Registration | |
|---|---|---|---|---|
| Brightfield Image | 24-bit color TIFF/BigTIFF or a JPEG | --image |
Supported | Supported# |
| 16-bit grayscale TIFF/BigTIFF or a JPEG | ||||
| Fluorescence Image | 8 or 16-bit grayscale single, multi-page TIFF/BigTIFF | --darkimage |
Not Supported (must use manual fiducial alignment in Loupe Browser) |
Supported# |
| 8 or 16-bit grayscale multiple single-page TIFF/BigTIFF or a JPEG | --darkimage |
|||
| 24-bit single colored image TIFF/BigTIFF or a JPEG | --colorizedimage |
|||
| CytAssist Captured Image | 24-bit color TIFF | --cytaimage |
Supported | Supported# |
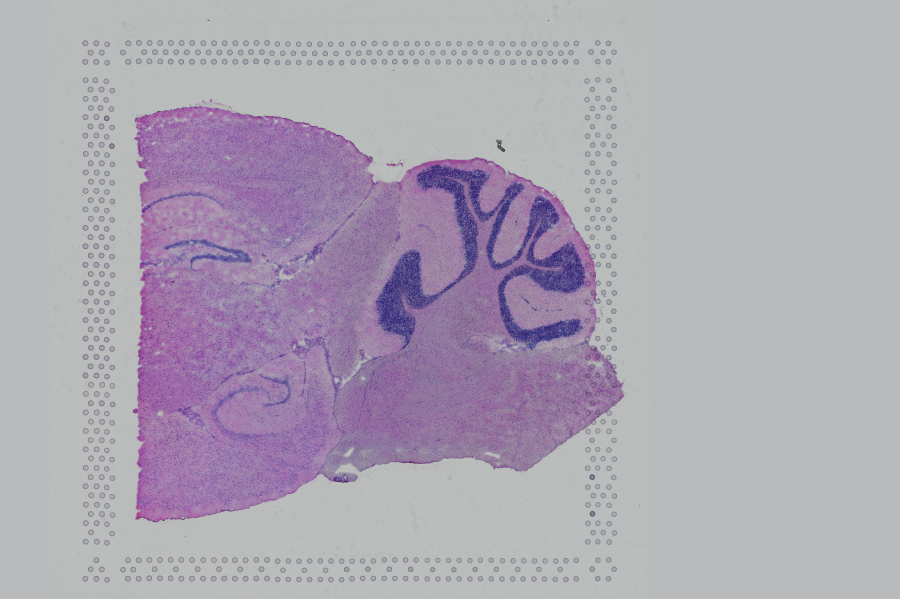
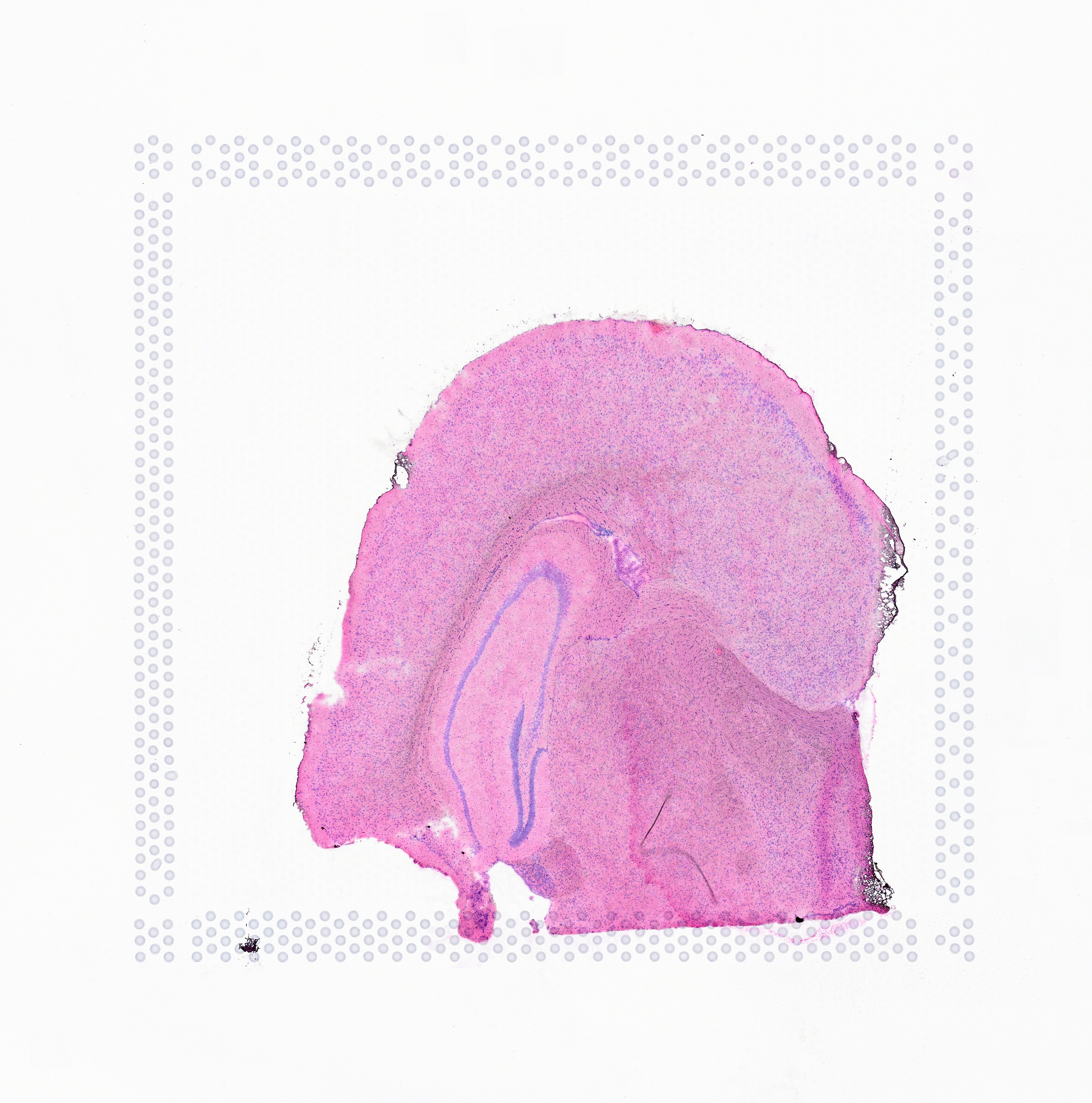
Space Ranger supports two styles of microscopy images: either a brightfield image stained with hematoxylin and eosin (H&E) or a fluorescence image comprising one or more acquisitions or channels (see below for examples of both, first row). The typical brightfield image is a transmission image, illuminated from below with white light and imaged from above. Dense areas of the sample, where tissue and cells lie, attenuate the transmitted light, producing an image that has a bright background and a dark or colored foreground. Fluorescence microscopy uses excitation with narrow band light and measures a signal emitted from either endogenous or added fluorophores, generating an image with a dark background and bright foreground. The fluorescence imaging demonstrated protocol for Visium is for immunofluorescence, where signal is generated by fluorophores conjugated to antibodies that bind to proteins of interest.
A fluorescence image comprises one or more grayscale intensity images (below, bottom row) and is often visualized as a single combined image with a different color assigned to each channel acquired (below, top-right). Each channel corresponds to a different fluorophore that binds to a different protein of interest. Space Ranger can take as input a brightfield image, one or more grayscale fluorescence channel images, or a color image made from combining and coloring fluorescence channel images in an external program such as ImageJ/Fiji or Photoshop. For technical details on imaging and examples of imaging artifacts that might lead to analysis errors, refer to the Visium Imaging Guidelines.
Space Ranger 2.0 added support for eosin stained CytAssist captured image, here by referred to as CytAssist image. The CytAssist workflow enables the use of a CytAssist image, which contains the fiducial frame for Gene Expression analysis, with the option to add a high resolution microscope image of the same tissue section in either brightfield and fluorescence image formats. For more details, refer to the CytAssist mediated Gene Expression analysis page.
 |
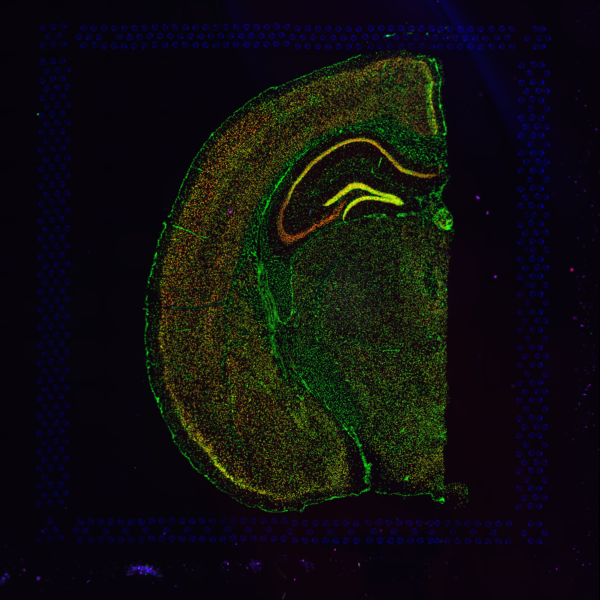 |
| Brightfield Image | Fluorescence Image Colorized |
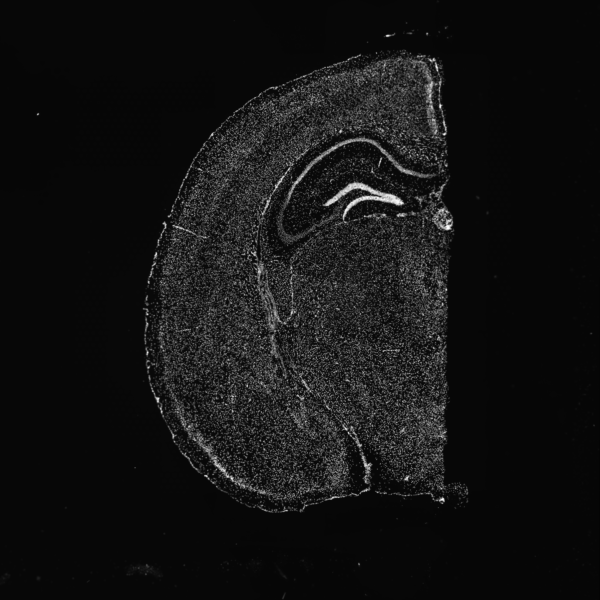 |
 |
 |
| Fluorescence Image, channel 1 | Fluorescence Image, channel 2 | Fluorescence Image, channel 3 |
 |
 |
| CytAssist Image, 6.5 mm Capture Area |
CytAssist Image, 11 mm Capture Area |
Depending on the slide type, Space Ranger requires images that meet minimum dimension requirements. In order to ensure uniform performance of the image processing in Space Ranger across a range of input resolutions, the input image is downsampled based on the slide version on which the tissue section was imaged. Space Ranger's automatic processing will not perform well on smaller images and they are explicitly rejected from processing. Note that any downsampling performed internally in Space Ranger does not impact visualization in Loupe Browser, which always uses the full resolution input image.
| Slide Version | Minimum Input Image Size | Space Ranger Downsampled Size ( tissue_hires_image.png) |
|---|---|---|
| V1 V2 |
2000 pixels in either dimension | 2000 pixels in either dimension |
| V4 | CytAssist image fixed at 3,000 pixels in both dimensions | 2000 pixels in both dimensions |
| V5 | CytAssist image only: fixed at 3,000 pixels in both dimensions | 2000 pixels in both dimensions |
| Standard Glass Slide (paired with CytAssist image) |
2,000 pixels in either dimension when used with a CytAssist image on V4 slide (Recommend 6,000 pixels or higher for accurate image registration |
2000 pixels in either dimension |
| 4000 pixels in either dimension when used with CytAssist image on V5 slide (Recommend 6000 pixels or higher for accurate image registration |
2,000 pixels in either dimension |
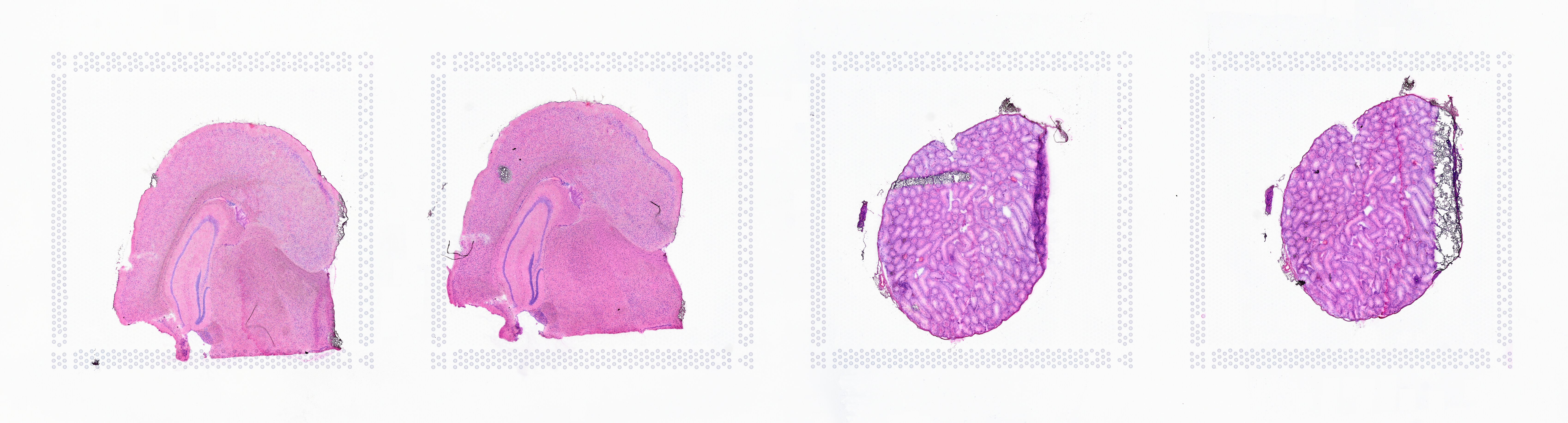
If the input image from Visium slide with 6.5 mm Capture Area (V1 or V2 ) has a great deal of whitespace to the left or right of the capture area, or the image includes a portion of the capture area either above or below the area being analyzed, the capture area in the input image may be downsampled too much in order to accommodate the extra white space. If the fiducial alignment or tissue detection results are not as expected for such images, 10x recommends cropping the images in order to remove the extraneous image area outside of the fiducial border. Images that are too tall or too wide can be easily cropped with freely available tools such as ImageJ/Fiji.
 |
 |
 |
| Too Wide | Too Tall | Correct |
The image supplied to Space Ranger must contain only one capture area from the Visium Gene Expression slide. The exact capture area of the supplied image is specified using the --area argument in the spaceranger count pipeline.
 |
 |
|||
| Incorrect Image | Correct | |||
The CytAssist captured image is fixed at 3,000 pixels in both dimensions and is an important check for Space Ranger to identify and process the inputs. Modifying the CytAssist image (e.g. cropping of white space, reducing file size etc) will make it incompatible as a pipeline input to Space Ranger. Use of a modified version of the CytAssist image will lead to errors when running spaceranger count. The CytAssist image dimension requirement also applies to Loupe Browser when using the manual image registration workflow.
Automatic image registration attempts to align the tissue section in the CytAssist image with the tissue section in the microscope image. In cases where there is more tissue in the microscope image than the CytAssist image, such as when the tissue section is larger than the CytAssist field of view, we recommend using the manual image registration workflow in Loupe Browser instead. Symmetrical tissue sections (e.g. circular punches) without any discernible morphological features may also initialize improperly during automatic image registration and require manual image registration.
Brightfield microscopy input images for use with Visium must be 24-bit color TIFF/BigTIFF, 16-bit grayscale TIFF/BigTIFF, or a JPEG.
Brightfield image input is specified in spaceranger count using the --image argument. In addition to these basic file type requirements for both Loupe Browser and Space Ranger,
the automatic image processing pipeline in Space Ranger can be used for brightfield images
that meet the additional restrictions outlined below. If these restrictions cannot be met, you can
still process your data using the manual alignment and tissue selection process in Loupe Browser.
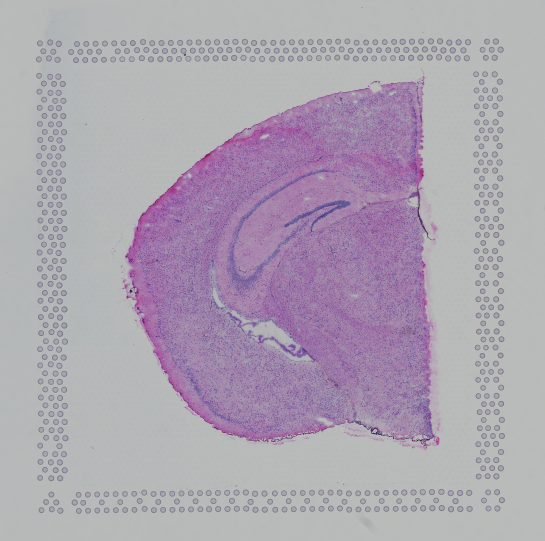
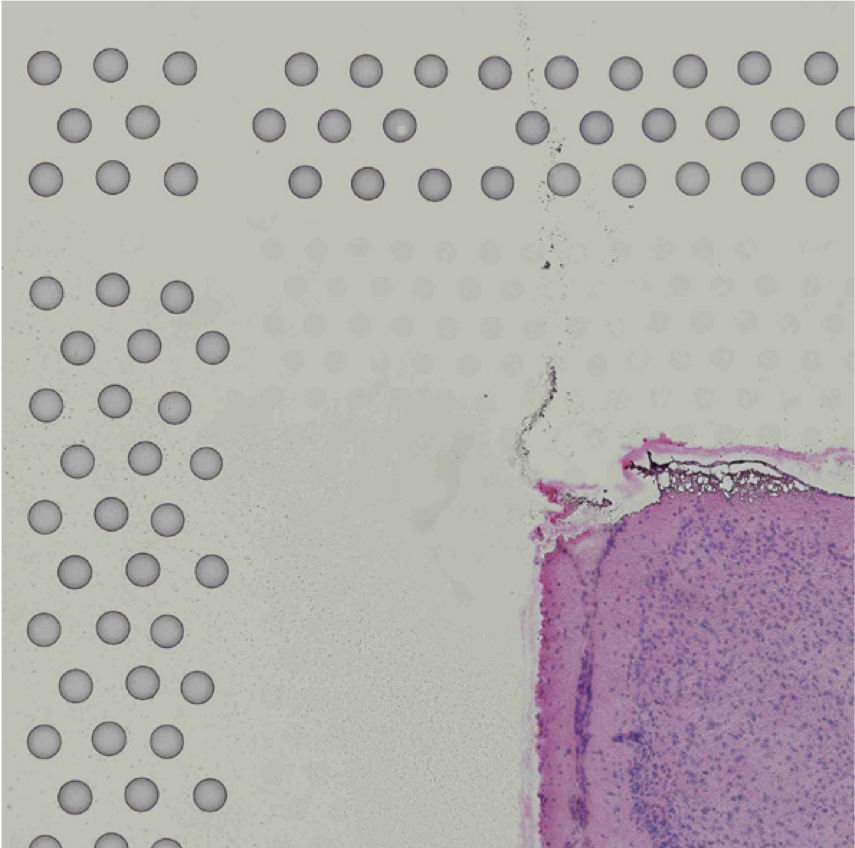
Images destined for automatic processing in Space Ranger should be oriented such that the hourglass fiducial marker is the in upper-left corner of the image. The image must be roughly axis-aligned, although slight rotations (e.g. less than 15 degrees) should be fine.
 |
 |
| Properly Oriented Slide Image | Corner with Hourglass Fiducial |
From Space Ranger 2.0 onwards, the pipeline will automatically rotate the fiducial frame and mirror the image to the expected orientation. This option can also be made explicit by using the --reorient-images=true flag supplied to spaceranger count. If this process fails, you can also correct for image rotation and mirroring either by using the manual
alignment step in Loupe Browser or by correcting the image using freely available tools such
as ImageJ/Fiji or Image Magick prior to processing with Space Ranger.
The automatic image processing in Space Ranger relies on the detection of spots in the fiducial frame. This process may perform poorly if the images are overexposed. Because the fiducial spots are slightly darker at the edges, very overexposed fiducials will appear as rings instead of solid circles. In general, poorly exposed images cannot be corrected after acquisition. If such images fail to process properly in the automatic pipeline, you can process such data through the manual alignment and tissue selection process in Loupe Browser prior to running Space Ranger.
 |
 |
| Properly Exposed Slide Image | Over-Exposed Slide Image |
Fluorescence images can be provided to the pipeline either as individual grayscale acquisitions or as a single, combined, color image. There are three possible input modes:
One or more acquisitions stored in a single, multi-page TIFF/BigTIFF file, where each page encodes an acquisition in
greyscale at 8 or 16-bit depth, up to six total pages. All pages must be the same dimensions in pixels and the same bit depth. This type of image input is specified in spaceranger count using the --darkimage argument.
One or more acquisitions each stored in individual, single-page greyscale 8 or 16-bit depth TIFF/BigTIFF files, or as JPEG files with up to six total files. All files must be the same size and bit depth. These types of image inputs are specified in spaceranger count by either providing the file names as a a comma separated list (--darkimage=channel_1.tiff,channel_2.tiff,channel_3.tiff,...) or by listing mutliple --darkimage arguments (--darkimage=channel_1.tiff --darkimage=channel_2.tiff ... ).
A color image where the user has colored and combined the individual acquisitions into a single image. This
format can be specified as either a 24-bit color TIFF/BigTIFF or a JPEG file. Note that with the options above,
Loupe Browser will allow you to combine and color the images during visualization. This option is provided for
users who wish to perform this function externally in software such as Photoshop or ImageJ/Fiji. This type of image input is specified in spaceranger count using the --colorizedimage argument.
Space Ranger will not accept images in other formats at this time, but 10x Genomics is open to feature requests for preferred image formats.
|
Processing of fluorescence microscope image requires fiducial alignment and tissue area selection using
manual alignment in Loupe Browser. The automatic registration and tissue detection analysis is not currently supported for fluorescence images in standard Gene Expression (GEX) analysis. The same constraint does not apply for CytAssist enabled GEX analysis with a fluorescence microscope image input. This is because the brightfield CytAssist image, which has the fiducial frame, is used for fiducial alignment and tissue detection and is compatible with the automatic image processing pipeline. |
Space Ranger 1.2.2 and Loupe Browser 5.0.1 introduced the ability to utilize TIFF images that contain multiple resolutions. This covers both pyramid format TIFF images that encode a progressive range of image resolutions, and images with a single, low-resolution preview image. This feature is implemented by identifying the highest resolution (largest number of pixels) in a single page within the file, then extracting all pages with the same shape (rows and columns). With this strategy in place, the other rules for TIFF files, as described above, remain in place and the software will accommodate file structures that include multiple fluorescence filter channels as well as a multi-resolution encoding. TIFF images where the highest resolution page is ambiguous (e.g., a file with one 6000x4000 page and a second 4000x6000 page) cannot be read.
Space Ranger 2.0.0 and Loupe 6.2.0 add support for QPTIFF images which are a type of pyramid TIFF, and a format used by whole slide imaging systems such as the Vectra Polaris™. Note that the QPTIFF image should correspond to a single Capture Area.