Cell Ranger6.1, printed on 03/30/2025
This page describes the cellranger multi output file structure, specifically for 5' Chromium Single Cell Gene Expression (GEX) and V(D)J data. Refer to the multi pipeline page to learn about running cellranger multi.
The output folder name is the same as the run ID you specified (e.g. sample345 used on the Running multi page). The subfolder named outs/ contains the main pipeline outputs:
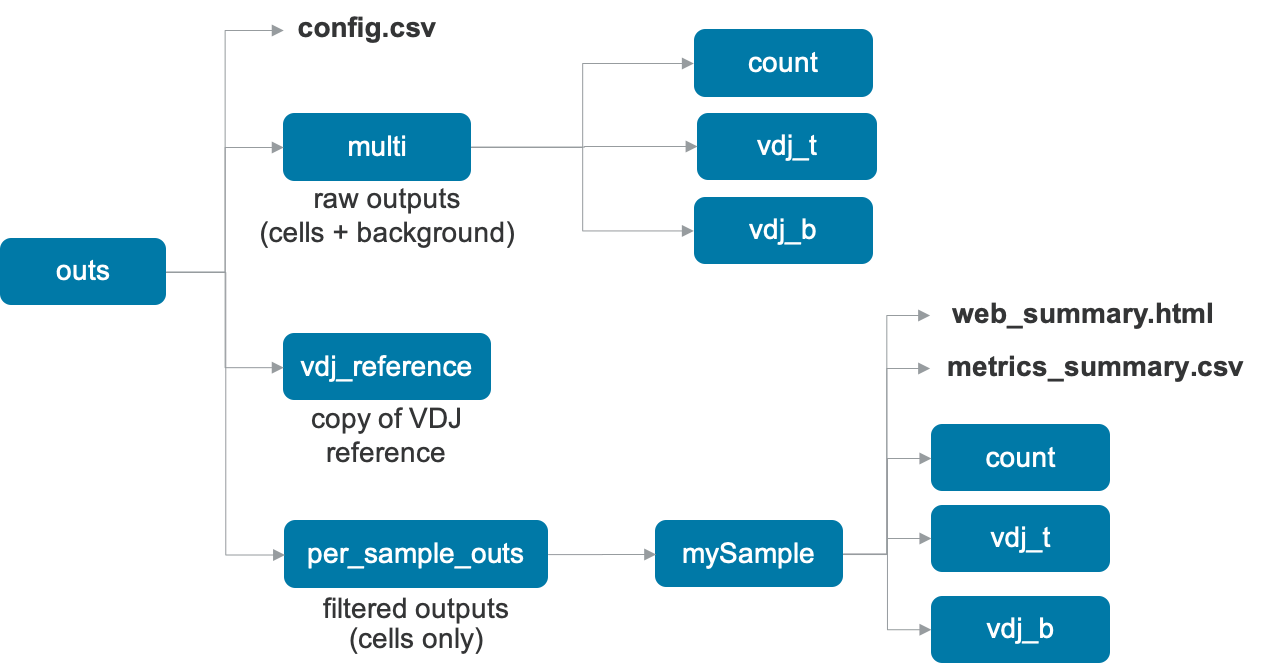
config.csv: a duplicate of the input config CSV file
vdj_reference/: folder containing a copy of the input V(D)J reference.
multi/: folder containing raw outputs, i.e., the data for all barcodes (GEMs) including cells and background:
| Name | Description |
|---|---|
count | Folder containing unfiltered results of 5' single cell gene expression analysis (cell-associated and background barcodes), described in the Raw Outputs section. |
vdj_t | Folder containing unfiltered V(D)J Immune Profiling analysis results for any T cells (cell-associated and background barcodes), described in the Raw Outputs section. |
vdj_b | Folder containing unfiltered V(D)J Immune Profiling analysis results for any B cells (cell-associated and background barcodes), described in the Raw Outputs section. |
| Name | Description |
|---|---|
web_summary.html | Run summary metrics and charts in HTML format, described in the Web Summary page. |
metrics_summary.csv | Run summary metrics file in CSV format, described in the Metrics page. |
count | Folder containing the results of any gene-expression and feature barcode analysis, similar to cellranger count, described in the Filtered Outputs section. Contains the feature-barcode matrix that can be used to load the data onto third party tools for downstream analysis. |
vdj_b | Folder containing the results of V(D)J Immune Profiling analysis for any B cells, similar to cellranger vdj, described in the Filtered Outputs section. |
vdj_t | Folder containing the results of V(D)J Immune Profiling analysis for any T cells, similar to cellranger vdj, described in the Filtered Outputs section. |