Cell Ranger ATAC2.0, printed on 04/02/2025
The Cell Ranger ATAC pipeline outputs a summary HTML file containing metrics and secondary analysis plots and results.
At the top of the summary HTML file we show the sample ID and its description followed by four key metrics that provide an overall view of the experiment. If any of the main metrics fall out of an expected range, a color-coded warning or alarm is displayed at the top of the page. The different sections of the summary HTML file are described next. Descriptions of metrics in the following sections can also be found by clicking the '?' next to the section header in the summary HTML file itself.
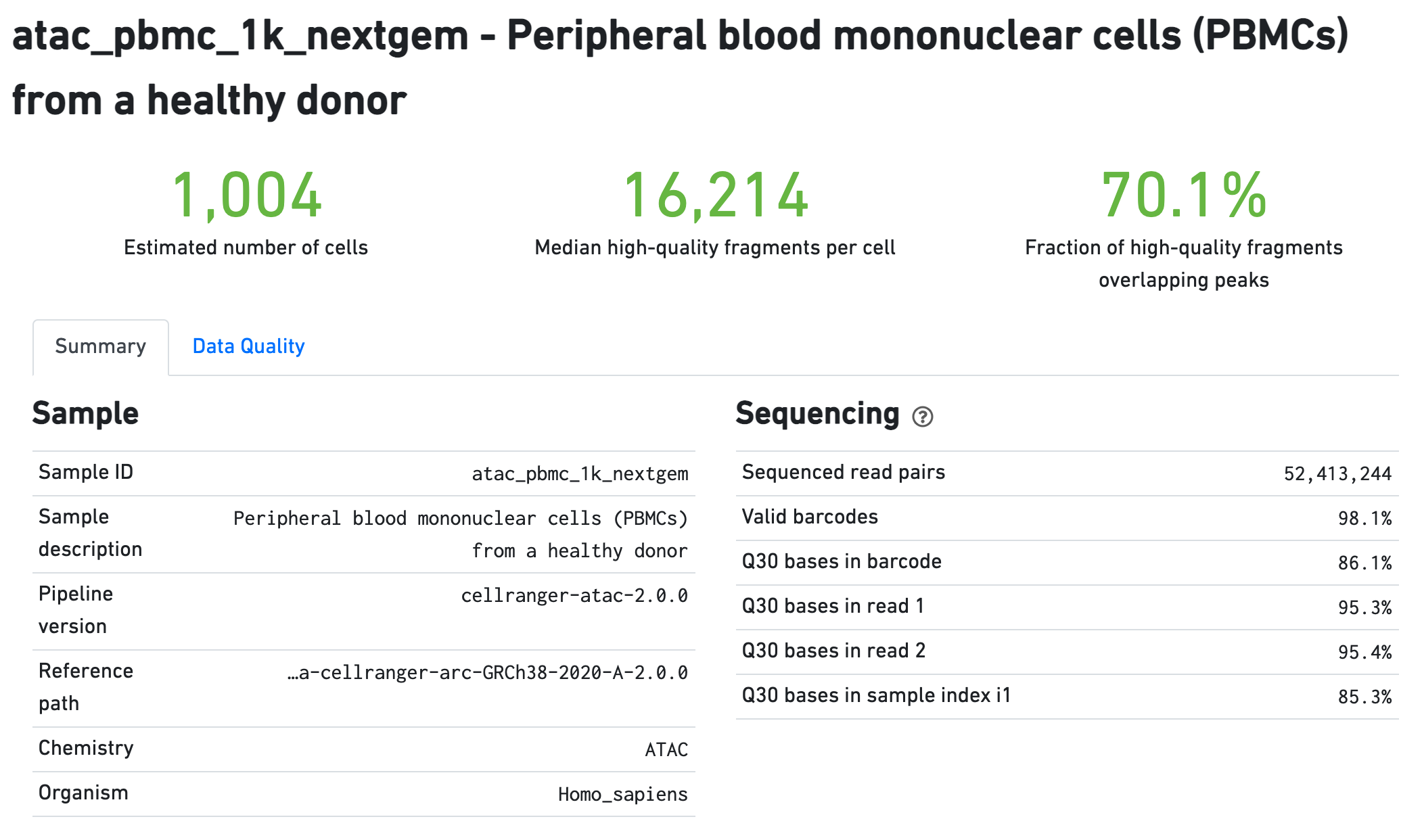
The Sample section summarizes the inputs to and version of the pipeline execution. Sequencing QC metrics are shown in the Sequencing section.
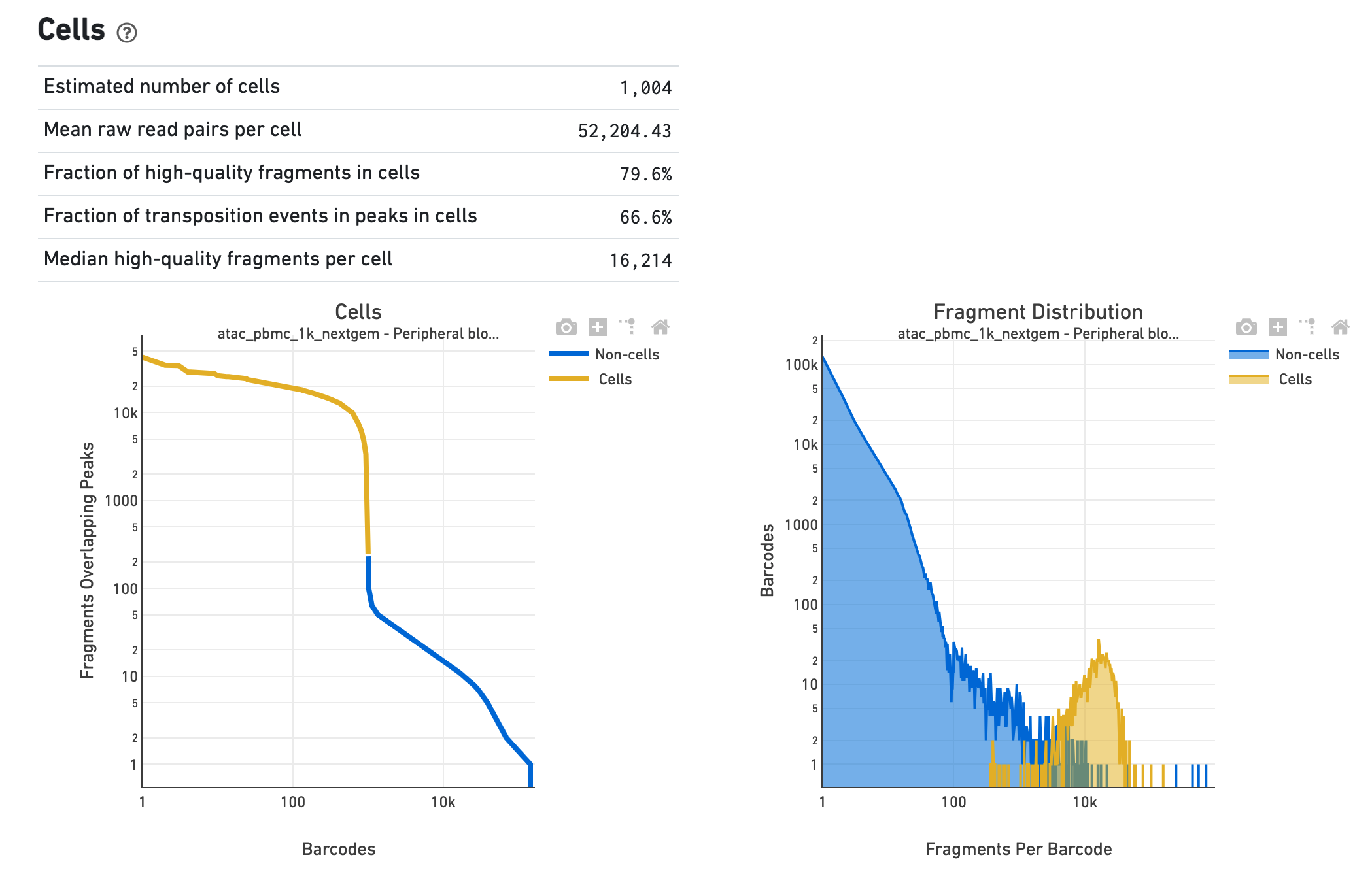
The Cells section shows metrics and plots related to identification of cells. We show the barcode rank plot (or knee plot) for fragments overlapping peaks (see Algorithms for details) and mark the barcodes that were inferred to be associated with cells. A steep drop-off is indicative of good separation between the cell-associated barcodes and the barcodes associated with empty droplets. We also show the distribution of the number of fragments per cell barcode for the non-cell and cell groups. Notice the knee plot is based on fragments that overlap peaks, whereas the distribution plot corresponds all fragments per barcode. The metrics in the table are key statistics derived from the data that help call cells on the knee plot and the histograms.
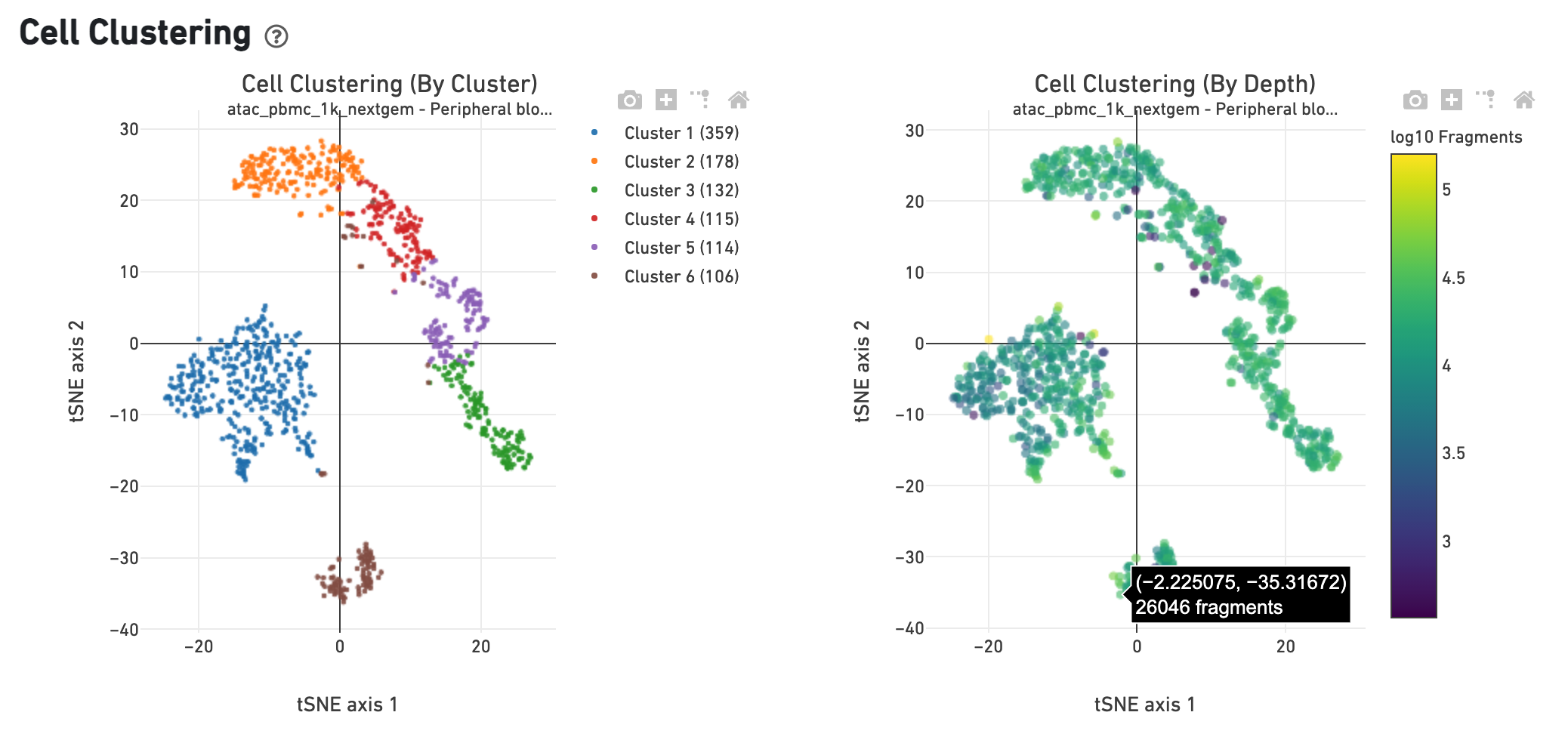
The Cell Clustering (By Cluster) plot shows the cell-associated barcodes in a 2-D t-SNE projection, with colors showing an automated graph clustering analysis which groups together cells that have similar chromatin accessibility profiles. In the Cell Clustering (Colored by Depth) plot, the same 2-D projection is shown, but cell coloring is based on the number of unique fragments associated with the barcode.
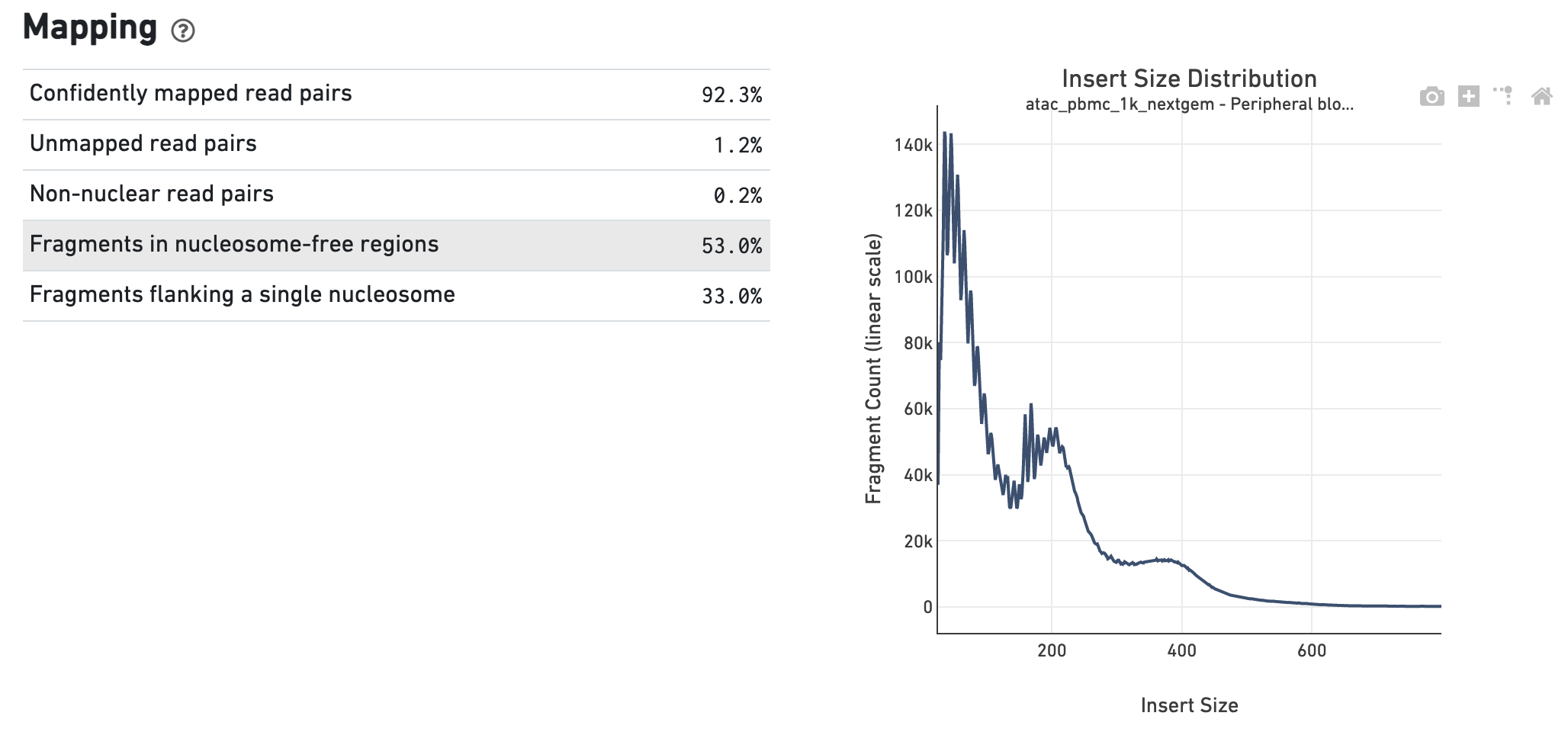
In the Insert Sizes section we show the Insert Size Distribution, and metrics derived from it. Single Cell ATAC read pairs produce detailed information about nucleosome packing and positioning. The fragment length distribution captures the nucleosome positioning periodicity.
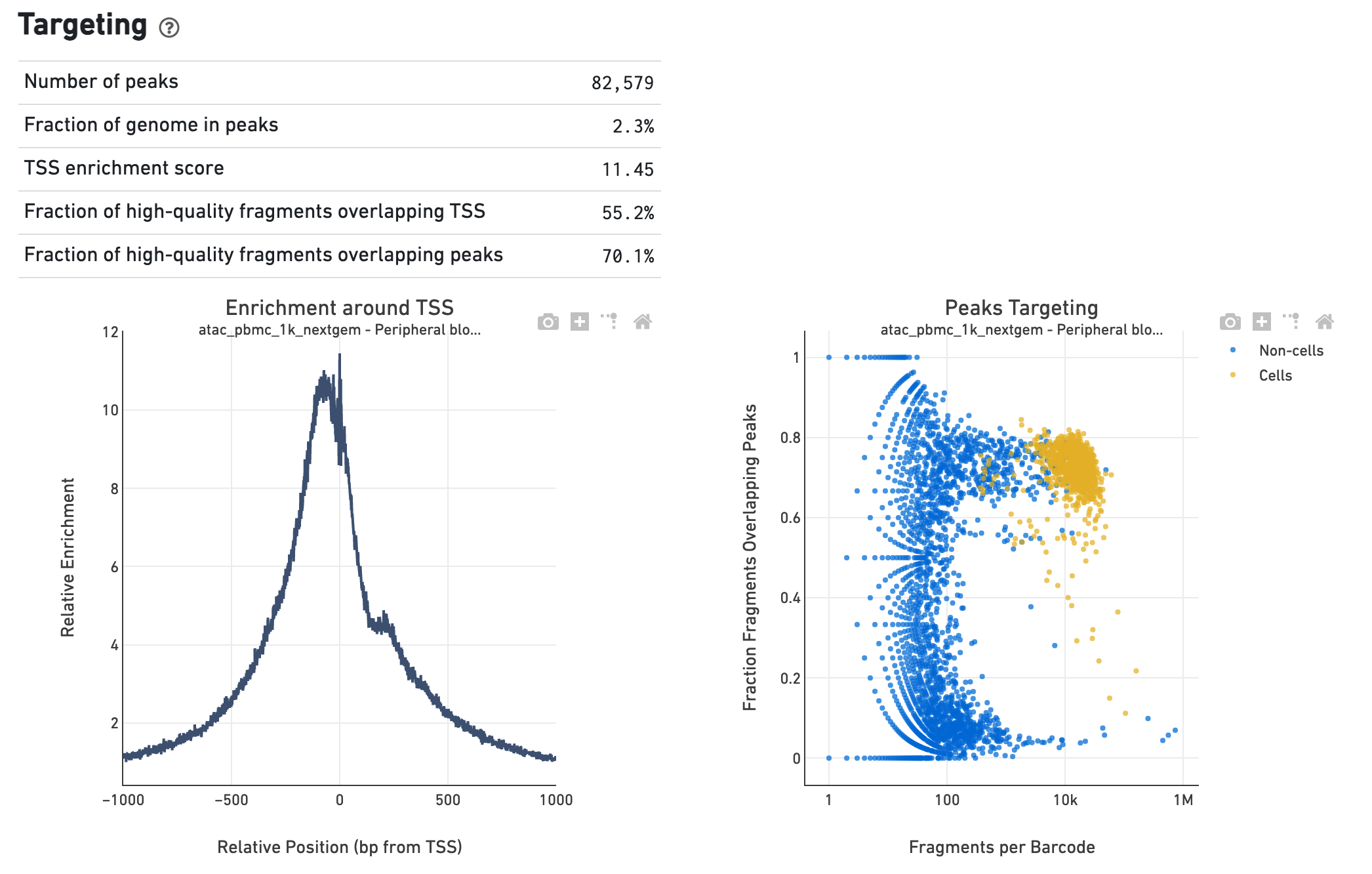
The Targeting section shows profiling of the chromatin accessibility behavior of the library at known, annotated, epigenetically relevant regions in the genome (see how the reference is built for more details). The first plot is the Transcription Start Site (TSS) profile, which is computed as the summed accessibility signal, or the number of cut sites per base, of all the barcodes irrespective of cell vs non-cell assignment, in a window of 2,000 bases around the full set of annotated TSSs and is normalized by the minimum signal in the window. This profile is helpful to assess the signal-to-noise ratio of the library, as it is well known that TSSs and the promoter regions around them have, on average, a high degree of chromatin accessibility compared to the intergenic and intronic regions of the genome. The "Enrichment score of transcription start sites" metric is derived from this profile. Note that this enrichment score depends on the source of tss sites as packaged with the reference. The second plot presents the variation in the number of on-target fragments, or fragments that overlap peaks, within each barcode group, namely cells and non-cells. For cell-associated barcodes it is expected that a high percentage of the barcode fragments overlap peaks. In the metrics table we summarize, for all the barcodes, the percentage that overlap not only peaks but also other kinds of annotated regions, such as enhancers and DNase hypersensitive sites.
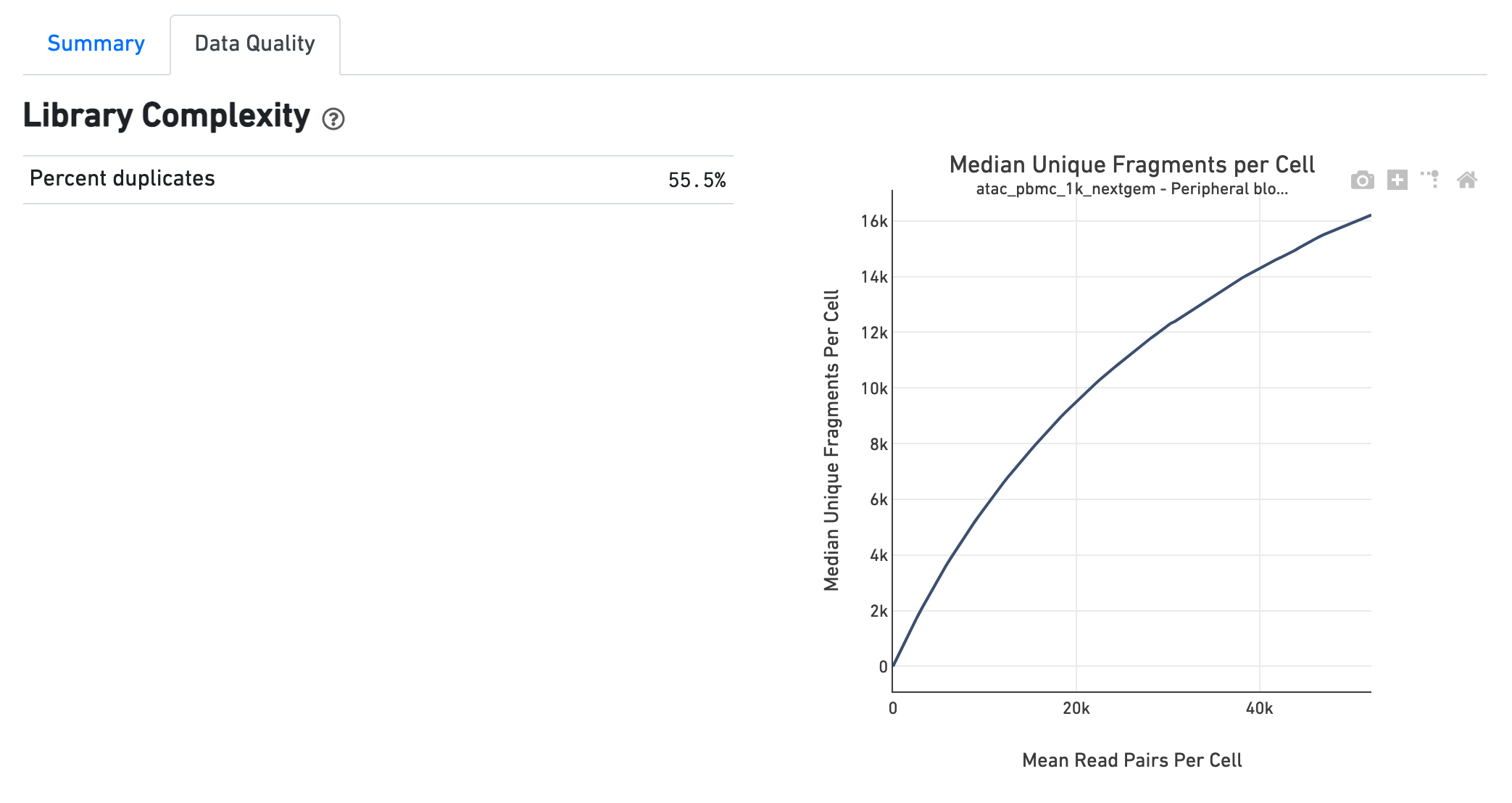
The Library Complexity section plots the observed per cell complexity, measured as median unique fragments per cell, as a function of mean reads per cell. The shape of the curve portrays the level of saturation in the library and can be used to inform decisions regarding sequencing depth to target for a sample. The metrics presented in the table summarize the complexity of the library measured for single cells and also in pseudo-bulk.
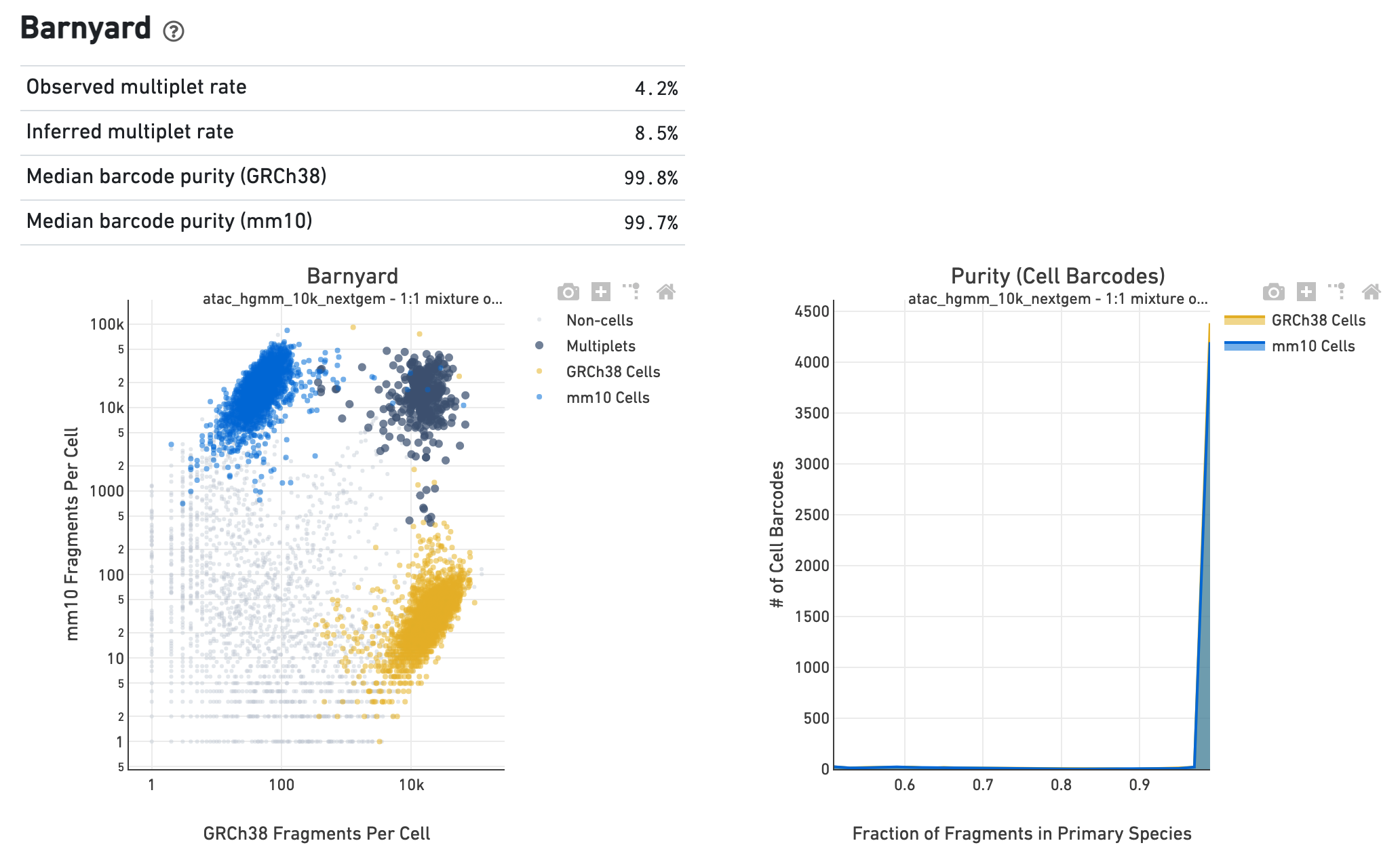
If the library corresponds to a multi-species experiment, the summary page will look different. A typical multi-species experiment, for instance, consists of mixing human and mouse cells. First, some of the metrics described before will have a version for each of the species in the experiment. Second, the summary will have a Barnyard section, which shows a scatter plot of all the barcodes and their number of associated fragments from each species, color coded by automatic assignment to non-cell, multiplet and cell barcode groups. It also shows, for each species, the distribution of barcode purity, measured as the fraction of fragments in a cell barcode that align uniquely to the species assigned to the barcode. The metrics in the table summarize key statistics from the data that generated the plots.
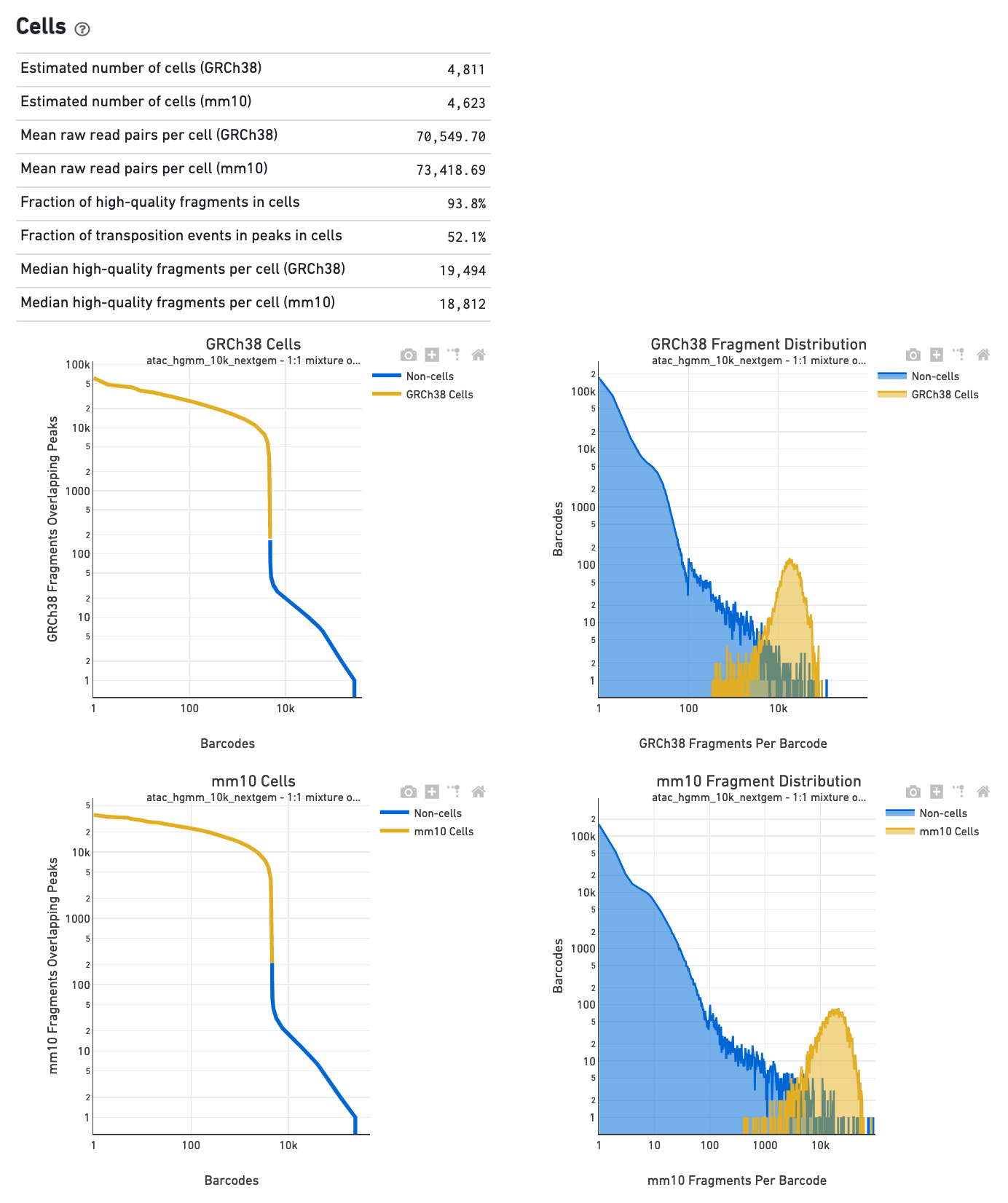
The Cells section in a multi-species library will show plots corresponding to each of the species.
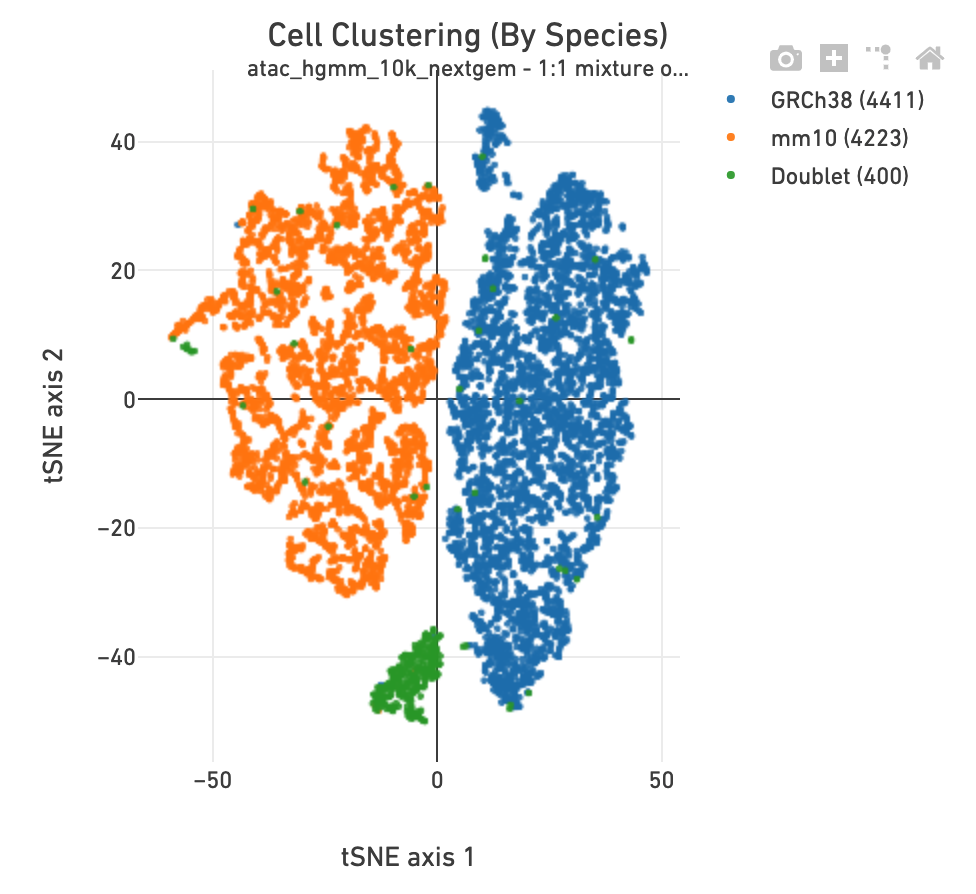
The Cell Clustering section in a multi-species library includes one more clustering plot, in which each barcode is colored according to its species assignment.