Cell Ranger ATAC6.4, printed on 03/29/2025
|
Documentation and downloads for 10x Genomics software products are moving to our redesigned software support site. This site is being maintained temporarily to support Cell Ranger ATAC, Cell Ranger ARC, and older versions of Cell Ranger, Space Ranger, Loupe Browser, and all versions of Loupe V(D)J Browser. |
The Loupe Browser ATAC tutorial demonstrates how to find significant features, analyze differential accessibility patterns, and explore substructure within a real-world dataset.
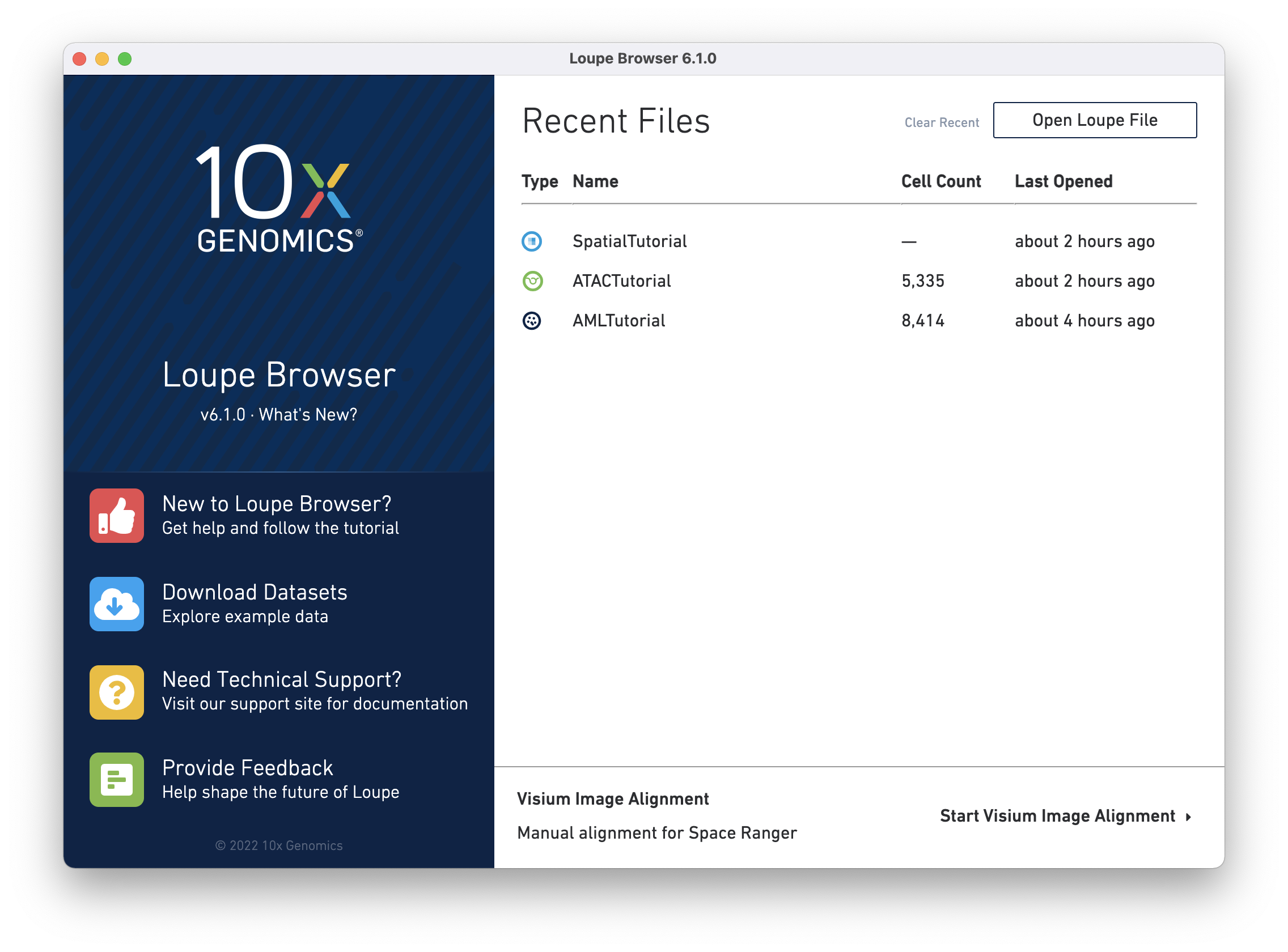
Before beginning the tutorial, make sure you have downloaded Loupe Browser. If this is your first time working with Loupe Browser, you can access the ATAC tutorial dataset by clicking on the ATACTutorial.cloupe link on the Recent Files page as seen below.
Once the file is loaded and you are familiar with some ATAC data concepts, click here to tour the user interface.
The ATAC tutorial dataset contains the results of a cellranger-atac run over a set of human peripheral blood mononuclear cells, with the standard Chromium™ Single Cell ATAC protocol.
The key metrics include:
Starting with Loupe Browser v6.1, users may choose to share anonymized non-biological usage data (i.e. no identifiable sample information such as files, cluster names, or any open text fields are tracked) to improve Loupe Browser performance. Sharing of usage data is optional, and users can change whether they want to share usage data any time. The user preference for usage data sharing will carry over within a major version (e.g. all 6.Y versions) but the user will be asked to confirm their preferences for each major version update.
In case you wish to change usage data sharing setting, click the Manage Usage Data from the Help menu and change the preference in the pop-up window.