Cell Ranger ATAC, printed on 03/29/2025
Cell Ranger ATAC pipelines run on Linux systems that meet these minimum requirements:
Note: Cell Ranger ATAC v2.0 was the last version that supported CentOS/RedHat 6.0 or Ubuntu 12.04.
In order to run in cluster mode, the cluster needs to meet these additional minimum requirements:
In order to run cellranger-atac mkfastq, the following software needs to be installed:
--jobmode=local by default, using 90% of
available memory and all available cores. To restrict resource usage, please
see the --localmem and --localcores flags for
cellranger-atac count at the link here for more information.| Limit | Recommendation |
|---|---|
| user open files | 16k |
| system max files | 10k per GB RAM available to Cell Ranger ATAC |
| user processes | 64 per core available to Cell Ranger ATAC |
The following data is based on time trials using Amazon EC2 instances, our PBMC 10k dataset, and version 2.0 of Cell Ranger ATAC. Performance is dependent on both the number of cells and the number of reads per cell. These plots will not be updated with every subsequent release of Cell Ranger ATAC, unless pipeline performance changes significantly.
| Instance | Threads | Memory (GB) | Core hours | Wall time | Storage HWM (GB) |
|---|---|---|---|---|---|
| r3.8xlarge (Ivy Bridge) | 32 | 64 | 129 | 5h 14m | 285 |
| m5.24xlarge (Skylake) | 96 | 384 | 92 | 1h 59m | 233 |
| r4.16xlarge (Broadwell) | 64 | 488 | 111 | 2h 58m | 233 |
| r1.32xlarge (Haswell) | 128 | 1952 | 137 | 2h 37m | 233 |
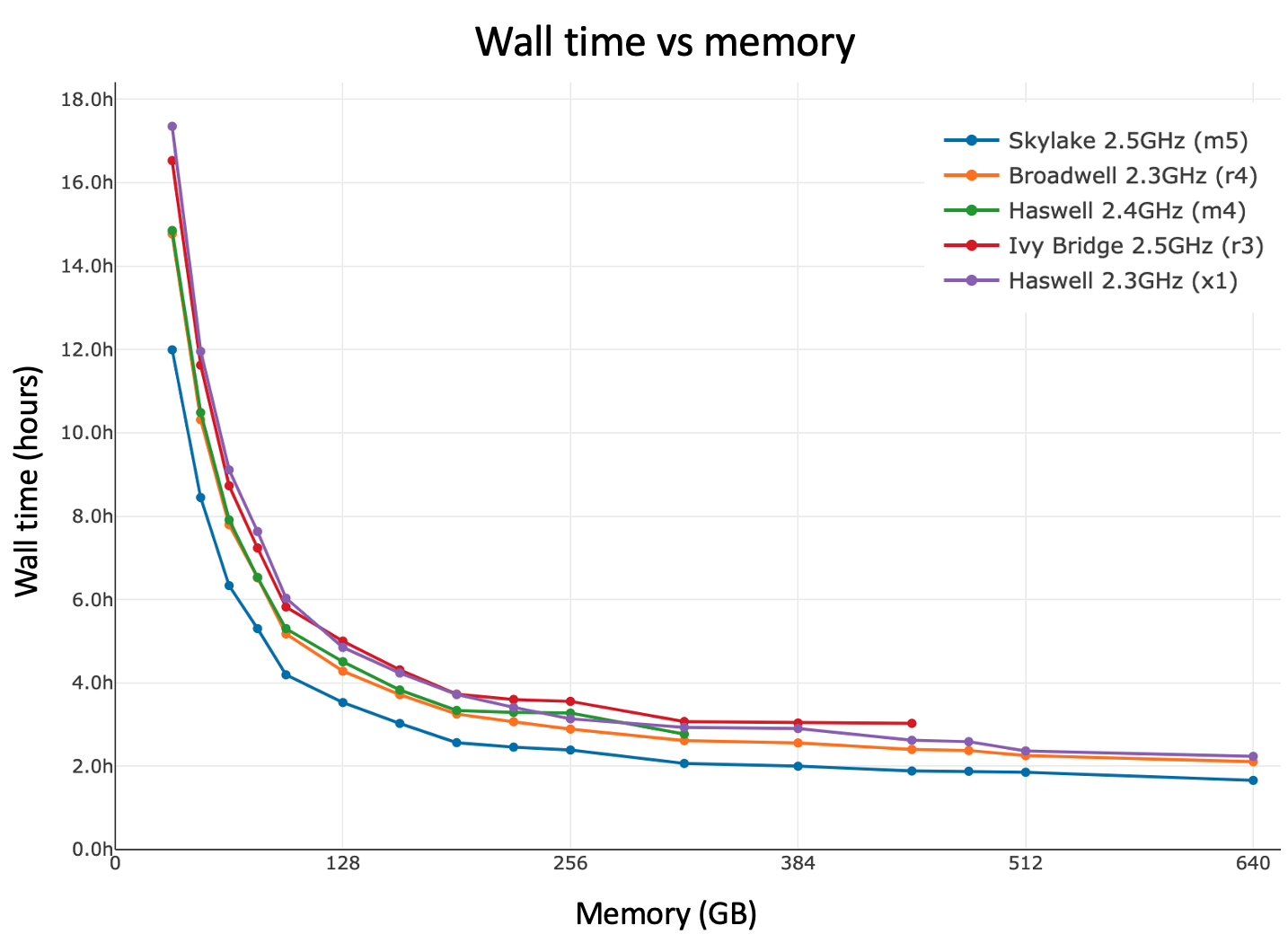
Here is cellranger-atac count walltime as a function of available memory for a variety of CPU architectures. In general, performance can be improved by allocating more than the minimum 64GB of memory to the pipeline. There is notable diminishing return beyond 160GB.
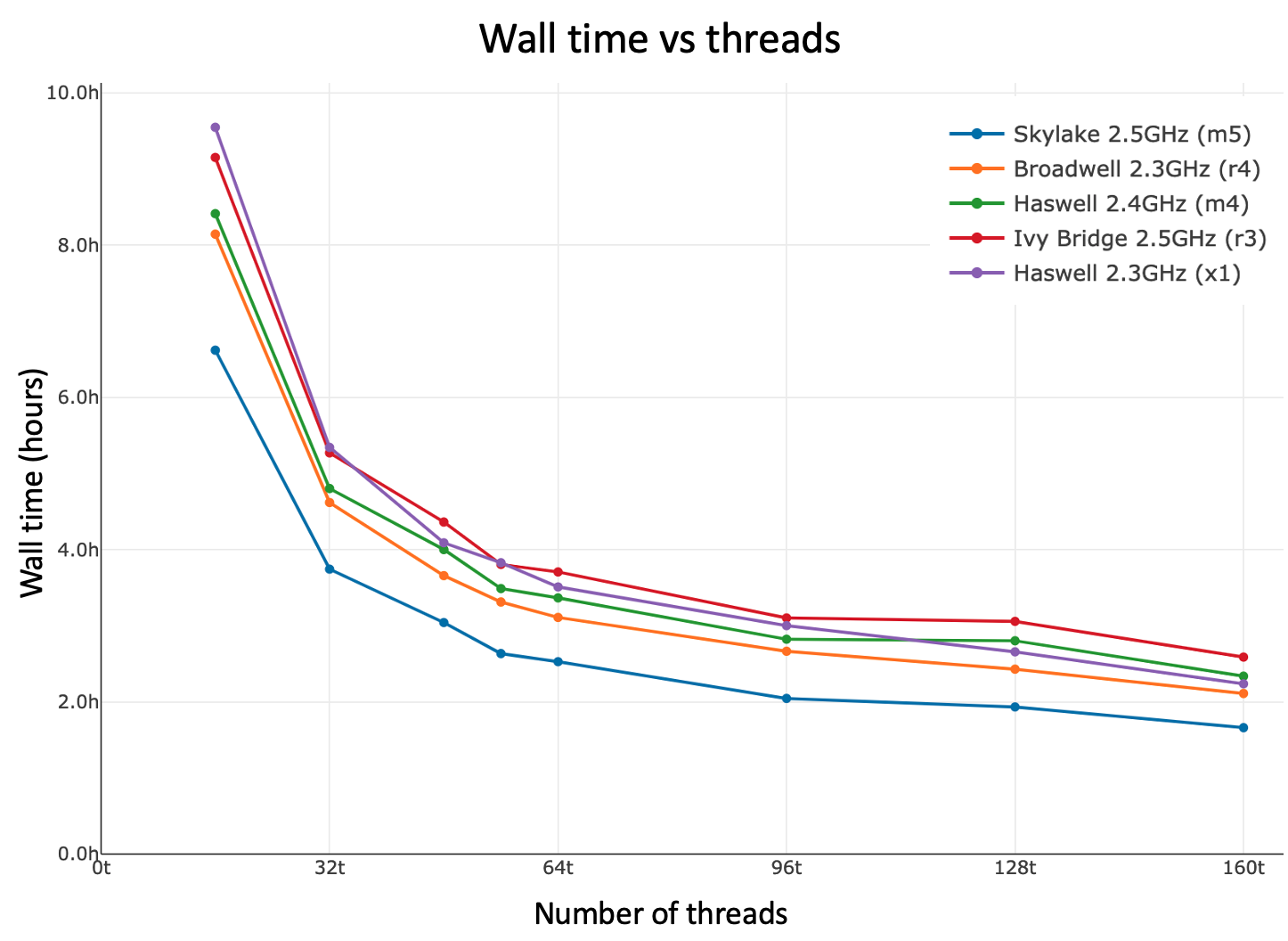
Shown below is cellranger-atac count walltime as a function of threads. If your system has ≫48 logical cores, you may want to run with --localcores=48 since there is diminishing return beyond 48 threads.