Cell Ranger ATAC6.4, printed on 03/29/2025
You can save your dataset workspace, including any custom clusters and feature lists by clicking on the Save (disk) icon on the toolbar.
If you prefer to create a new version of a .cloupe file, choose Save As from the File menu, or type →→ (Windows) or →→ (Mac). You'll be prompted to create a new .cloupe file somewhere in your file system.
Loupe Browser files are self contained. To share a dataset with collaborators, send them the .cloupe file.
There are a variety of mechanisms to export data and graphics from your ATAC dataset. The toolbar, Categories List, Feature Table, Filter List, Feature Table and Peak Viewer all have export functionality.
If you find that the exported screenshot of the plot has points that are too small or large, you can manually adjust the point size. To do so, disable marker auto-scaling and modify the point scale percentage.
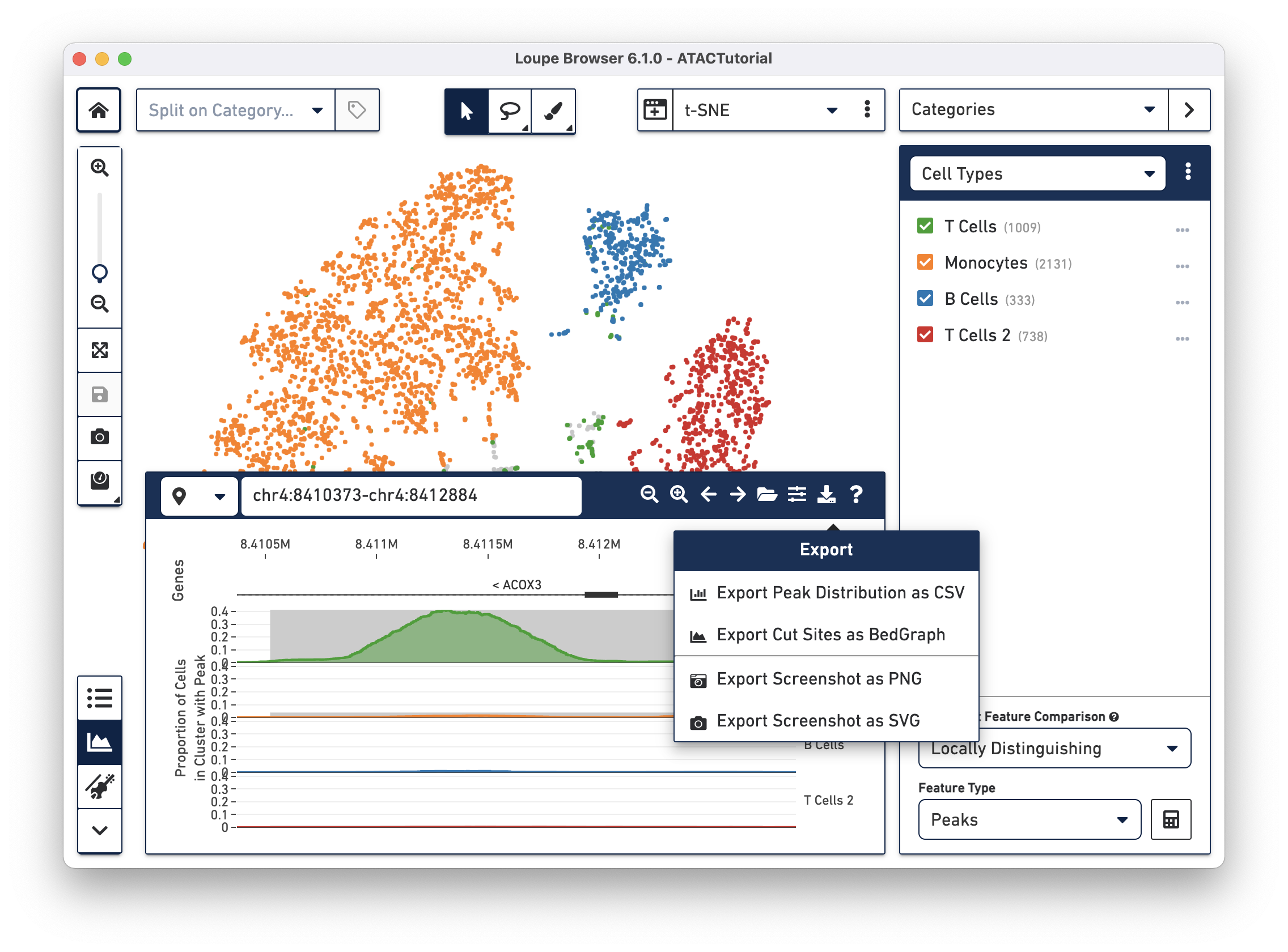
This concludes the Loupe Browser ATAC tutorial. Read about how to generate your own .cloupe files with the Cell Ranger ATAC pipeline.
If an error occurs in the program, send a bug report by clicking on Generate Bug Report from the Help menu. A .tar.gz file containing logs from your most recent Loupe Browser session is created. Send that .tar.gz file to support@10xgenomics.com. Add the subject line Loupe Browser Error to your message.
You can also submit general feedback and feature requests to support@10xgenomics.com.
We hope this tutorial has made you familiar with the capabilities of Loupe Browser, and made you excited to process your own data. We hope you find it to be the easiest, fastest and most enjoyable way to interpret your single cell ATAC data.