Cell Ranger7.0, printed on 02/01/2025
While most of the antibody-related metrics and counts are computed in parallel with their gene expression counterparts, there are some unique aspects of protein libraries that require specific analysis steps in Cell Ranger.
| Note: The same Cell Ranger algorithm is used to process Antibody Capture (Single Cell Gene Expression, Single Cell Immune Profiling) and Antigen Associated Capture (Single Cell Immune Profiling) libraries. Please note that Antigen Associated Capture library construction and analysis in the absence of Gene Expression and Antibody Capture is not recommended. |
Protein aggregates in antibody staining experiments cause a few GEMs to have extremely high UMI counts. There is a two-step process to detect and filter out such GEMS.
First, as part of the UMI counting pipeline, Cell Ranger looks for pairs of UMIs that differ by only one base (i.e., are Hamming distance one apart) and implements a UMI correction step, during which these reads will be combined into a single UMI count by correcting the UMI with fewer reads into the UMI with more reads. While such correction events are typically rare, we observed that the corrections rates are sometimes abnormally high in Antibody Capture libraries, a phenomenon that always correlates with extremely high UMI counts. These high UMI counts will cause saturation of the UMI space, leading to false UMI corrections. Protein aggregates are one leading cause of such high UMI counts and correction rates.
The plot below shows a particularly bad example of protein aggregation, where a handful of barcodes accounted for almost 77% of all reads, with extremely high correction rates in these reads. Currently, we consider a barcode an aggregate if it has more than 10K reads, 50% of which were corrected.
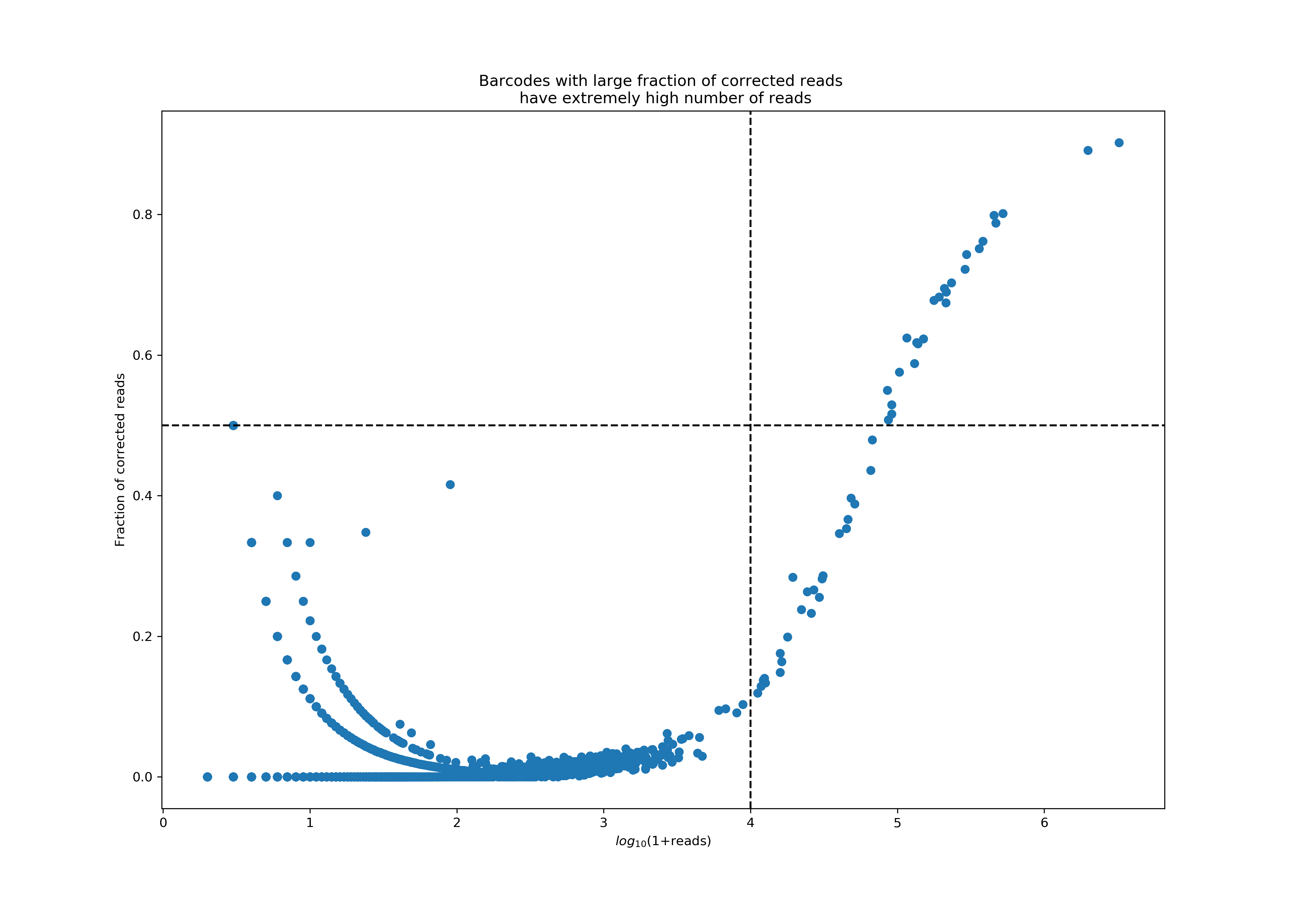
Second, in addition to tracking UMI correction events and using the high correction rate as a flag for protein aggregation, Cell Ranger directly uses protein counts to deduce aggregation events. The key insight is that large antibody panels are typically used to stain a wide diversity of cell types, and seeing high counts of many unrelated proteins in a GEM is a sign that such a GEM contains protein aggregates.
This algorithm activates only if five or more antibodies (or antigens, or dextramers, or other features specified as "Antibody Capture" in the provided feature reference) with at least 1,000 counts are detected. Using the total number of such features, Cell Ranger automatically decides which percentage of those need to be detected on a GEM for it to be considered further. Next, if in such GEMs the required number of antibodies exceed their pre-defined thresholds for high counts (currently defined as being in the 25 highest counts across all GEMs), they will be flagged as protein aggregates.
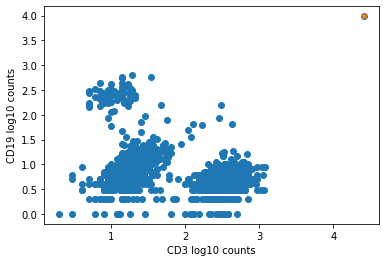
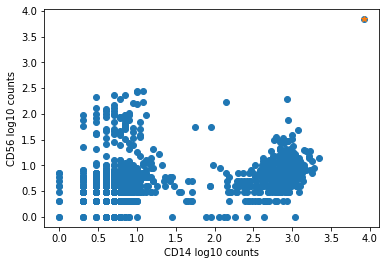
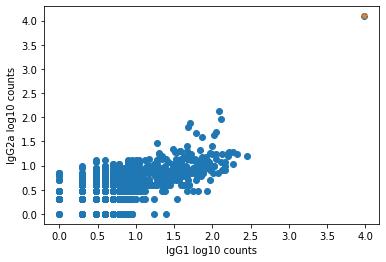
The plot below shows an example of such an aggregate barcode. Completely unrelated antibodies such as CD3, CD19, CD14, CD56, and even mouse isotype controls IgG1 and IgG2a are enriched in the barcode marked in orange (top right corner), which the pipeline flagged as an aggregate.
 |
 |
 |
Cell Ranger combines the aggregate barcodes from both steps above and removes them from the final feature-barcode matrix, and reports the fraction of reads associated with such barcodes in the "Antibody: Fraction Reads in Aggregate Barcodes" metric on the web summary. In addition, two more related metrics are available in metrics_summary.json; "Antibody: Number of Aggregate Barcodes" is the number of detected aggregate barcodes, and "Antibody: Fraction Reads that have Corrected UMIs" is the fraction of all reads that have a correction event.
In order to reduce the gene expression matrix to its most important features, Cell Ranger uses Principal Components Analysis (PCA) to change the dimensionality of the dataset from (cells x genes) to (cells x M) where M is a user-selectable number of principal components (via num_principal_comps). The pipeline uses a python implementation of the IRLBA algorithm, (Baglama & Reichel, 2005), which we modified to reduce memory consumption. For samples where Antibody Capture is an input library, the pipeline will slice out the feature counts from the full matrix and perform a PCA on these log-transformed antibody counts, $log_{2}\text{(count + 1)}$.
Since cell surface proteins offer a unique and complementary view of the cell types on top of the genes they express, Cell Ranger runs a popular t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm to visualize the protein counts in 2-D space. For samples where Antibody Capture is an input library, the pipeline will slice out the feature counts from the full matrix and perform a t-SNE on these log-transformed antibody counts, $log_{2}\text{(count + 1)}$ (unlike the gene expression part of the feature-barcode matrix, where t-SNE is run on the PCA-reduced space from raw counts). These t-SNE projections can then be visualized with Loupe Browser versions 3.0 and later.
Below, the left panel shows a traditional t-SNE with gene counts only, overlaying the counts of CD8a antibody. The middle panel shows CD8a protein expression overlaid on t-SNE projections computed on antibody counts only. The right panel shows the expression of CD8A gene on the antibody-derived t-SNE projections for comparison.
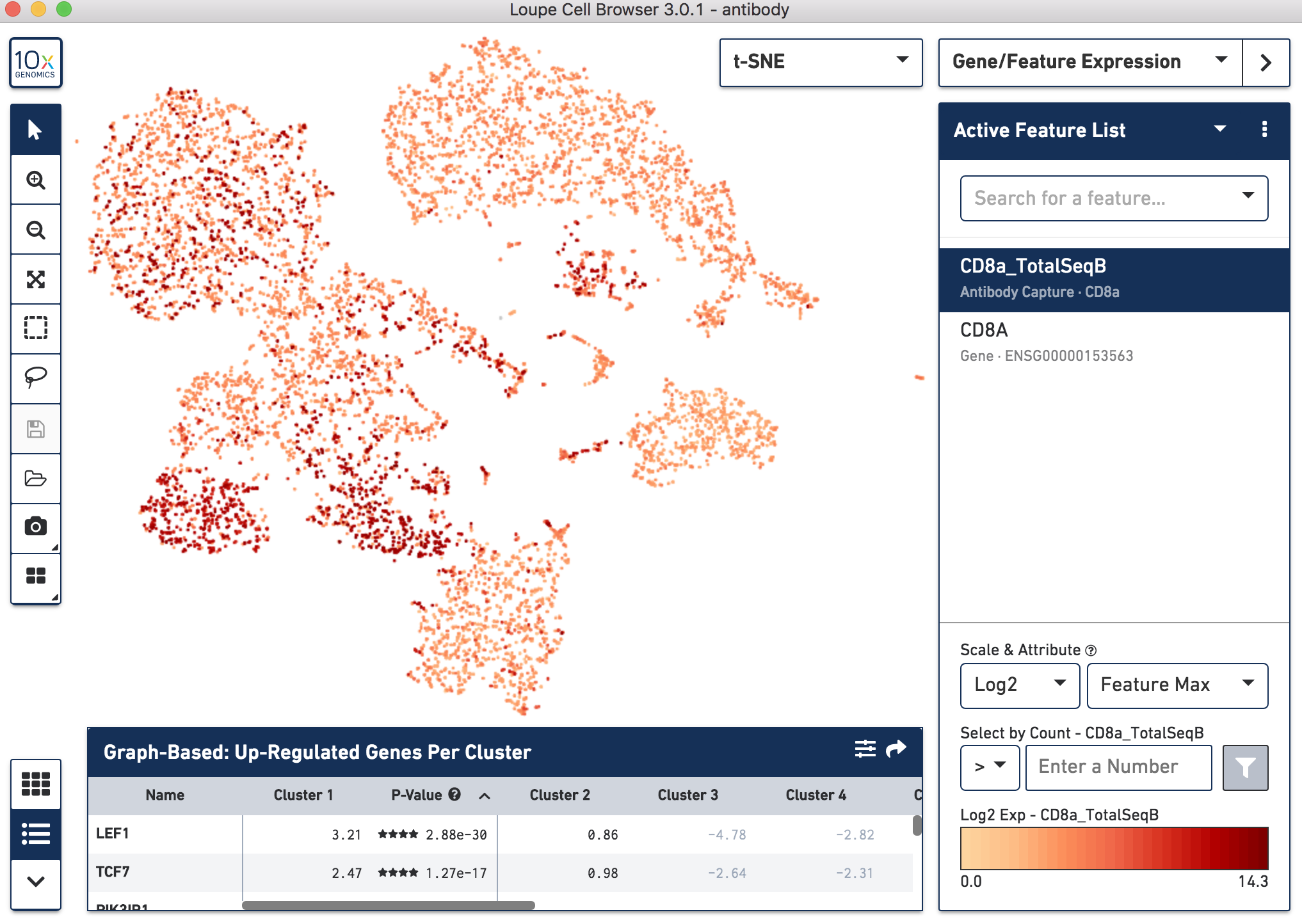 |
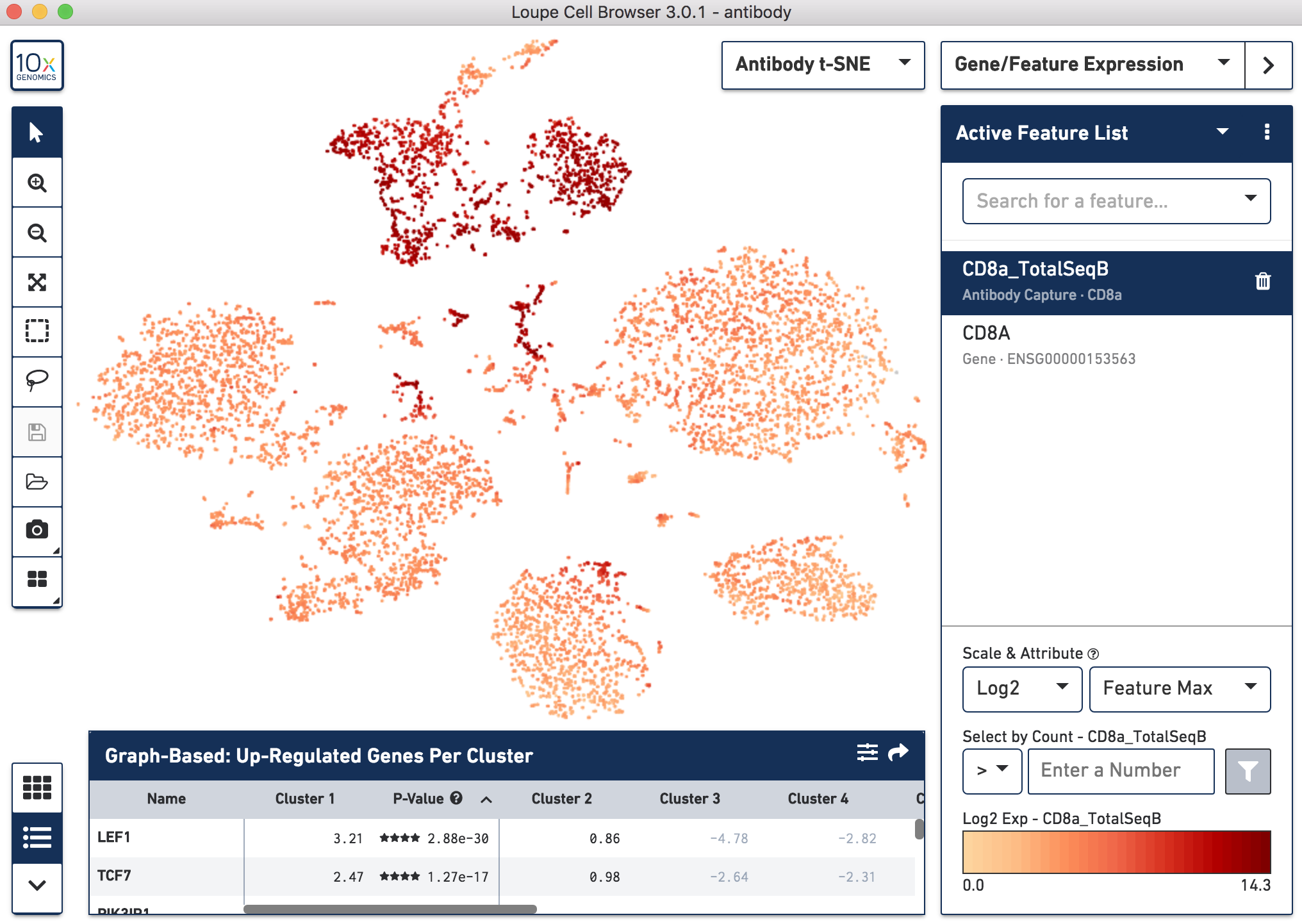 |
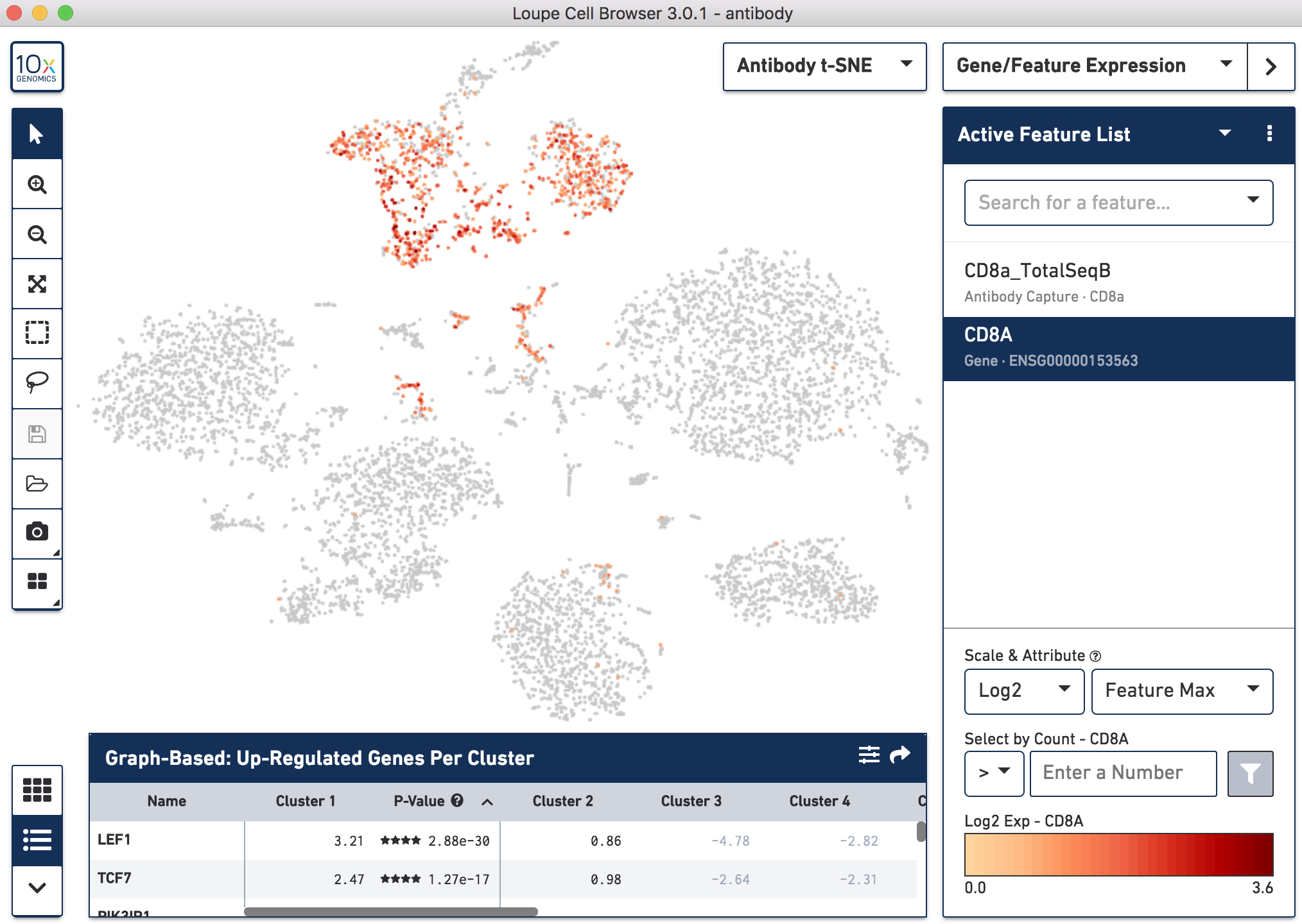 |
Cell Ranger also supports visualization with UMAP (Uniform Manifold Approximation and Projection), which estimates a topology of the high dimensional data and uses this information to estimate a low dimensional embedding that preserves relationships present in the data (McInnes et al, 2018). The pipeline uses the python implementation of this algorithm by McInnes et al (2018). UMAP coordinates are available in the pipeline output analysis/umap directory. Similar to t-SNE plots, in samples with Antibody Capture reads, the pipeline will slice out these feature counts from the full feature-barcode matrix and perform UMAP on log-transformed antibody counts, $log_{2}\text{(count + 1)}$.