Space Ranger1.0, printed on 03/26/2025
Multiplexed data from some Illumina platform sequencing instruments may exhibit a slightly elevated rate of "index hopping" - a phenomenon where samples sequenced together can "swap" sample indices, causing some data to be misassigned between samples.
In order to avoid this issue in the Visium assay, Space Ranger requires "dual-indexing," where multiplexed libraries are generated with both i7 and i5 sample indices. Sequencing this additional sample index uses available cycles (bases) in a given Illumina sequencing kit and may reduce the number of cycles available to capture transcript bases in read 2. The commonly used 100 cycle kit allows for a 75 base read 2 (R2), and the 150 cycle kit allows for a 120 base R2. These read lengths may not lead to optimal performance.
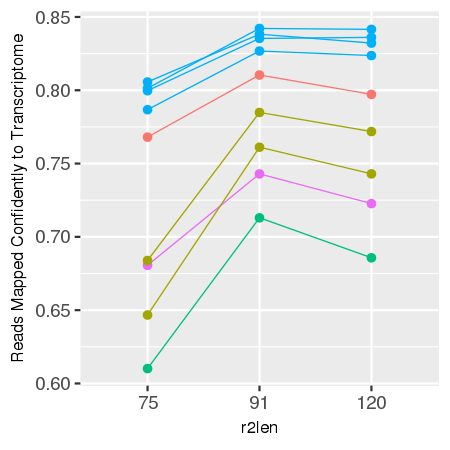
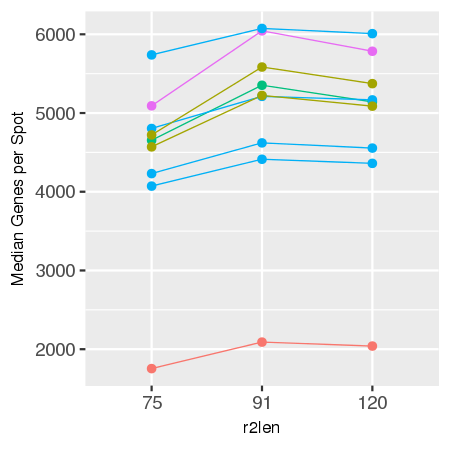
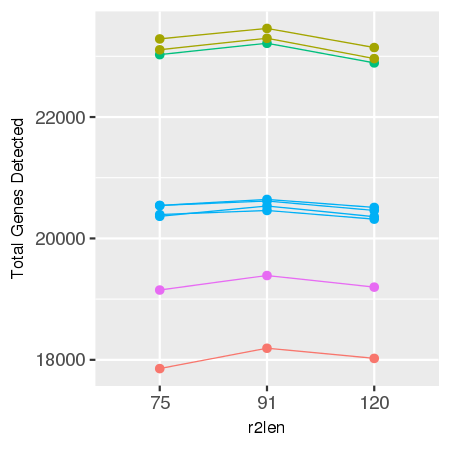
Gene expression analysis, such as in Space Ranger, relies on mapping reads to a known transcriptome and genome, and this process is affected by the length of R2. In our experiments, optimal mapping rates, and therefore sensitivity, are achieved with an R2 length of 91 bases, with less optimal performance seen for shorter or longer reads.
If you've sequenced with more than 91 bases, Space Ranger can trim
your data, if you desire, allowing you to find an optimum for your
sample. See the --r2-length option to spaceranger
count. If you've sequenced shorter reads, the
analysis will still work, but you may see reduced sensitivity. A
comparison of 75, 91, and 120 cycle read 2s for some sample data
data is shown below for reference.
 |
 |
 |
 |
| Sensitivity by R2 Length for Sample Tissue Types | |
All input tissue images for use with Visium must be 24-bit color TIFF, 16-bit grayscale TIFF, or JPEG. In addition to these basic file type requirements for both Loupe Browser and Space Ranger, the automatic image processing pipeline in Space Ranger imposes additional restrictions outlined below. If these restrictions cannot be met, you can still process your data using the manual alignment and tissue selection process in Loupe Browser.
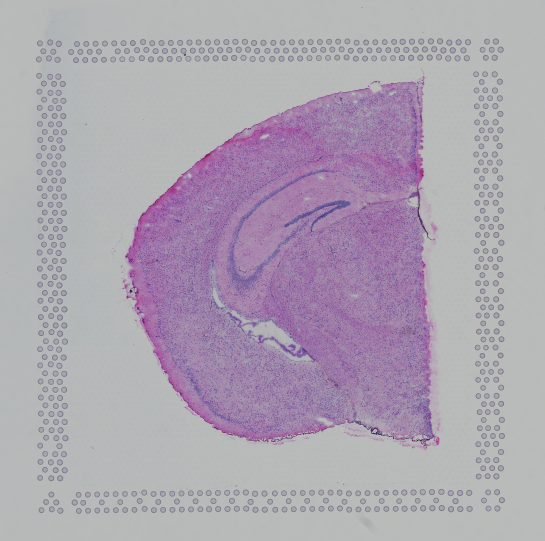
Images destined for automatic processing in Space Ranger must be oriented such that the hourglass glyph is the in upper-left corner of the image. The image should be roughly axis-aligned, although slight rotations (e.g. less than 15 degrees) should be fine.
 |
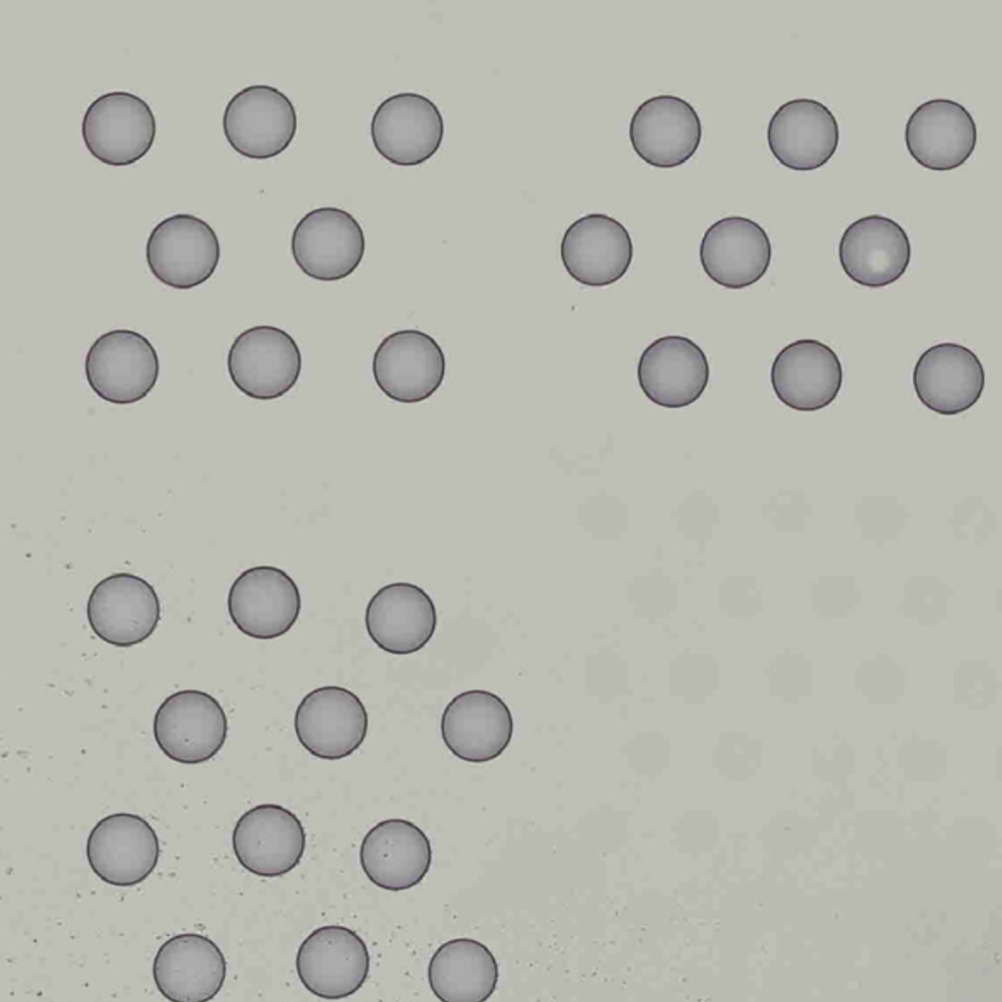 |
| Properly Oriented Slide Image | Corner with Hourglass Fiducial |
Note that image rotation (or mirroring) can be easily corrected with freely available tools such as ImageJ/Fiji or Image Magick.
In order to ensure uniform performance of the automated image processing across a range of input resolutions, the input image is downsampled to be no larger than 2000 pixels in either dimension. Space Ranger's automatic processing will not perform well on images smaller than 2000 pixels in each dimension. Note that downsampling does not impact visualization in Loupe Browser, which uses the full resolution input image.
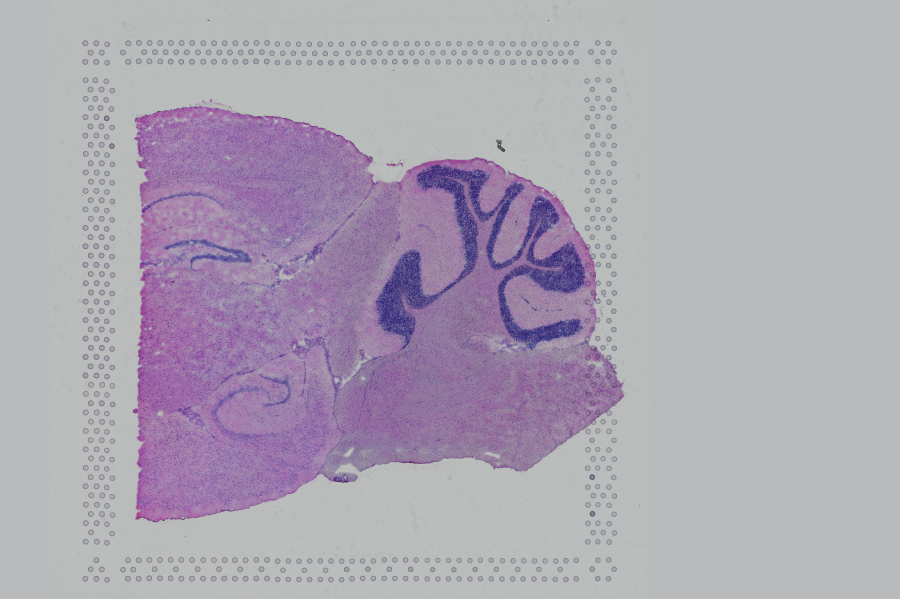
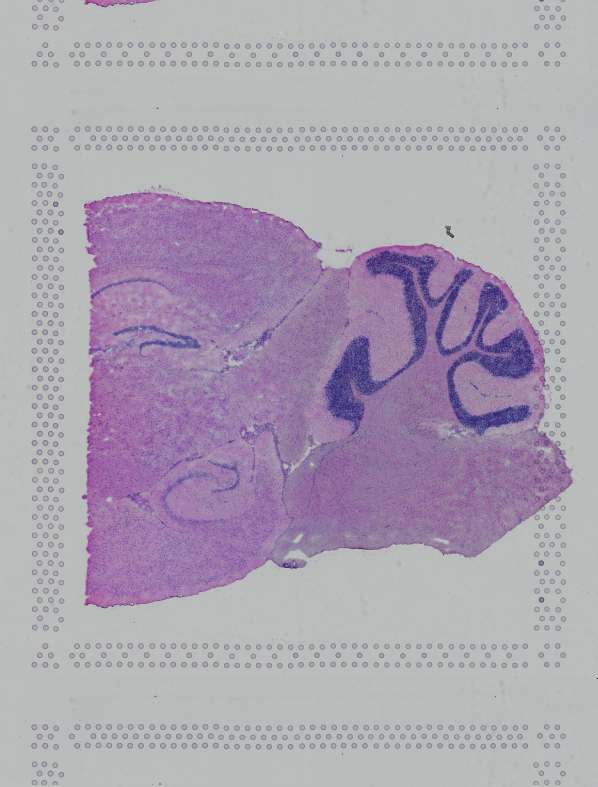
If the input image has a greal deal of whitespace to the left or right of the capture area, or the image includes a portion of the capture area either above or below the area being analyzed, the capture area in the input image may be downsampled too much in order to accommodate the extra white space. If the fiducial alignment or tissue detection results are not as expected for such images, we recommend cropping the images in order to remove the extraneous image area outside of the fiducial border.
 |
 |
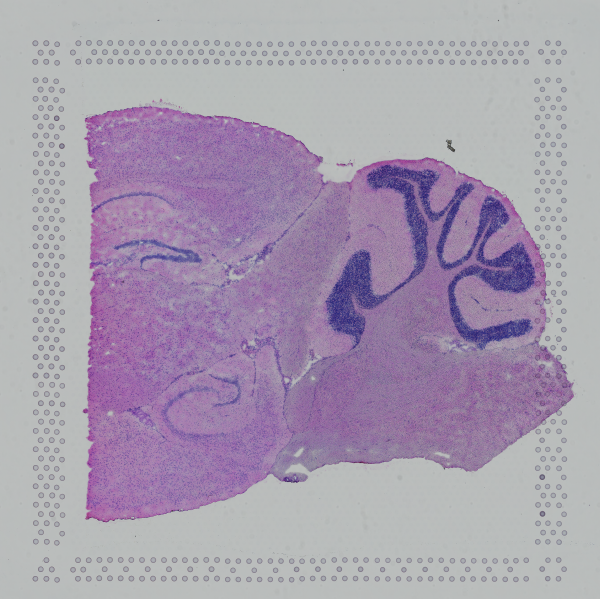 |
| Too Wide | Too Tall | Correct |
Images that are too tall or too wide can be easily cropped with freely available tools such as ImageJ/Fiji.
The automatic image processing in Space Ranger relies on the detection of spots in the fiducial frame. This process may perform poorly if the images are overexposed. Because the fiducial spots are slightly darker at the edges, very overexposed fiducials will appear as rings instead of solid circles. In general, poorly exposed images cannot be corrected after acquisition. If such images fail to process properly in the automatic pipeline, you can process such data through the manual alignment and tissue selection process in Loupe Browser prior to running Space Ranger.
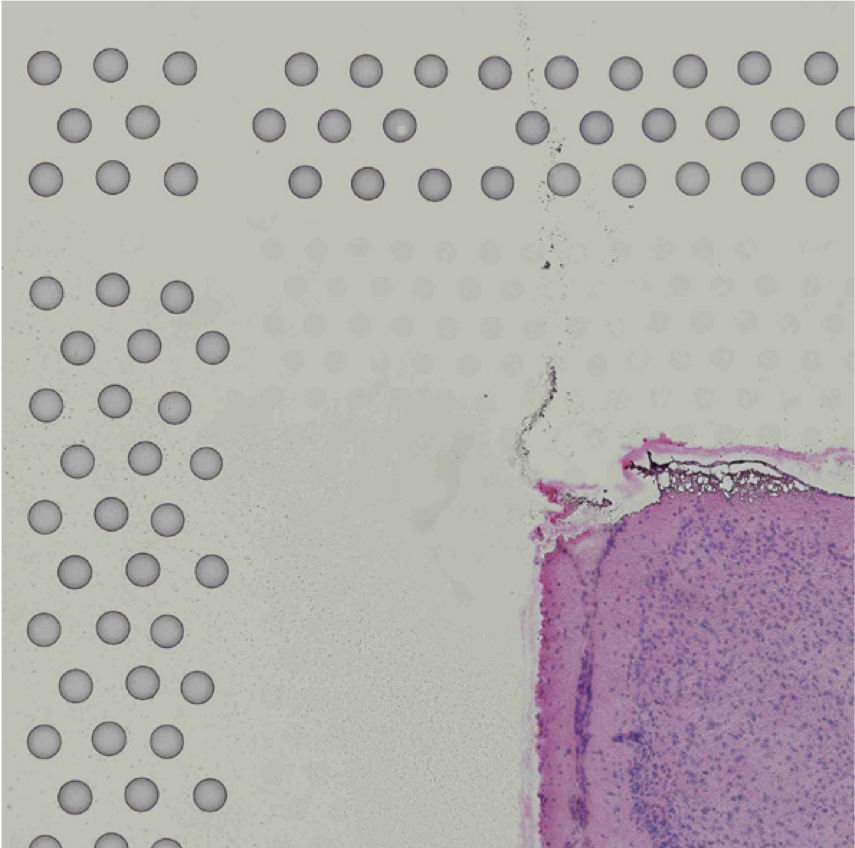 |
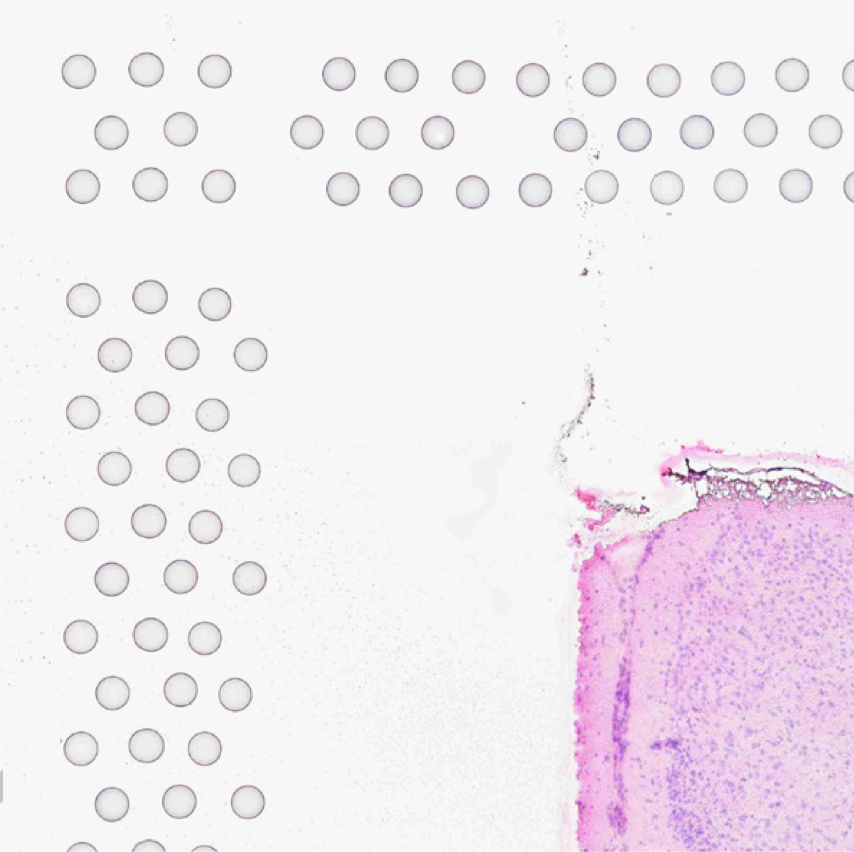 |
| Properly Exposed Slide Image | Over-Exposed Slide Image |