Space Ranger1.0, printed on 03/26/2025
Space Ranger relies on image processing algorithms to solve two key problems with respect to the slide image: deciding where tissue has been placed and aligning the printed fiducial spot pattern. Tissue detection is needed to identify which capture spots, and therefore which barcodes, will be used for analysis. Fiducial alignment is needed so Space Ranger can know where in the image an individual barcoded spot resides, since each user may set a slightly different field of view when imaging the Visium capture area.
These problems are made difficult by the fact that the image scale and size are unknown, an unknown subset of the fiducial frame may be covered by tissue or debris, each tissue has its own unique appearance, and the image exposure may vary from run to run.
If either of the procedures described here fail to perform adequately, the user may use the manual alignment and tissue selection wizard in Loupe Browser. Note that in most cases, the pipeline will not be able to indicate that a failure has occurred and the user is encouraged to check the quality-control image on the front page of the Web Summary that Space Ranger outputs.
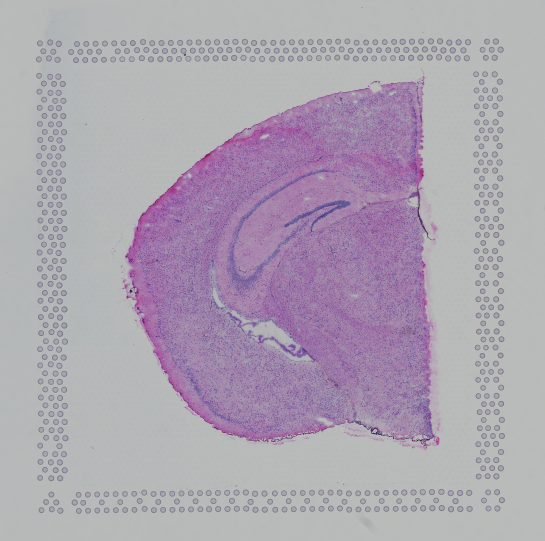
Finally, in order to use the automatic analysis described here, the image must be oriented with the "hourglass" fiducial in the upper-left corner:
 |
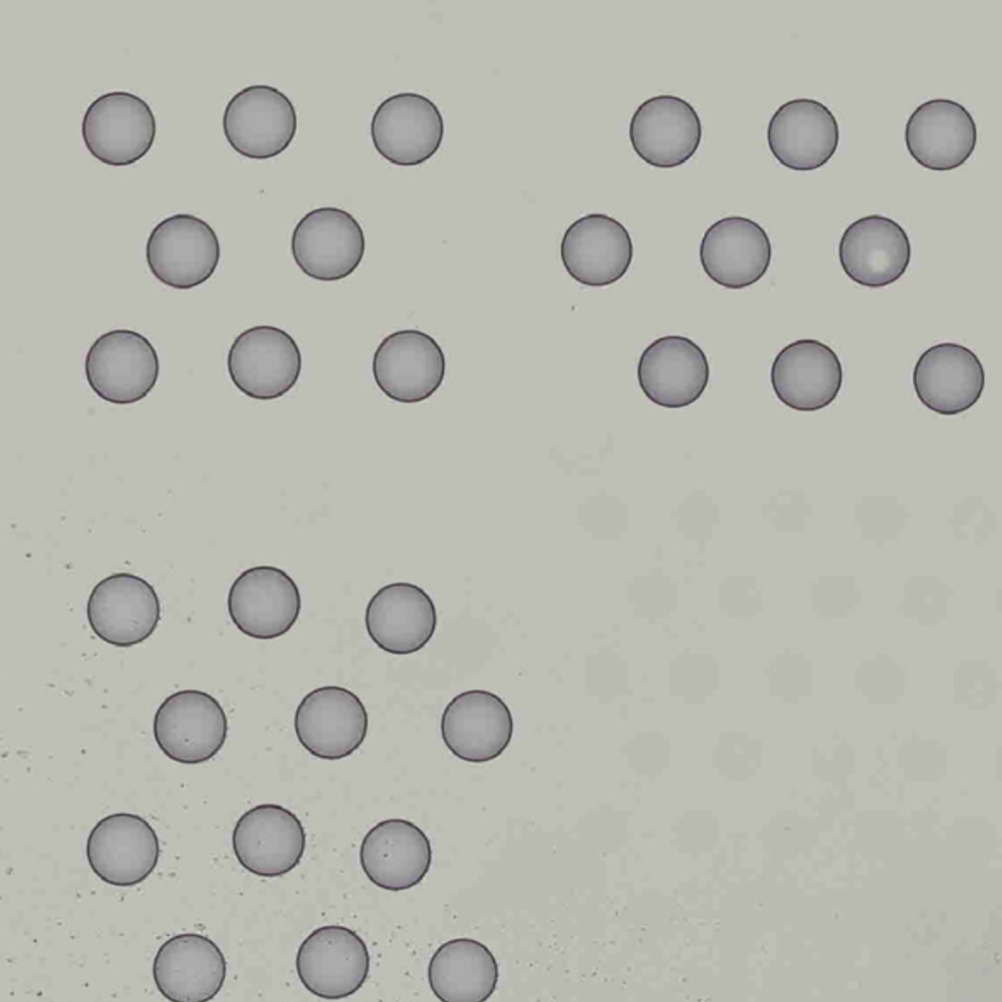 |
| Properly Oriented Slide Image | Corner with Hourglass Fiducial |
Visium contains a system for identifying the slide-specific pattern of invisible capture spots printed on each slide and how these relate to the visible fiducial spots that form a frame around each capture area. The fiducial frame has unique corners and sides that the software attempts to identify.
This process first extracts features that "look" like fiducial spots and then attempts to align these candidate fiducial spots to the known fiducial spot pattern. The spots extracted from the image will necessarily contain some misses, for instance in places where the fiducial spots were covered by tissue, and some false positives, such as where debris on the slide or tissue features may look like fiducial spots.
After extraction of putative fiducial spots from the image, this pattern is aligned to the known fiducial spot pattern in a manner that is robust to a reasonable number of false positives and false negatives. The output of this process is a coordinate transform that relates the Visium barcoded spot pattern to the user's tissue image.
Each area in the Visium slide contains a grid of 4,992 capture spots populated with spatially barcoded probes for capturing poly-adenylated mRNA. In practice, only a fraction of these spots are covered by tissue. In order to restrict Space Ranger's analysis to only those spots where tissue was placed, Space Ranger uses an algorithm to identify tissue in the input brightfield image.
Using a grayscale, downsampled version of the input image, multiple estimates of tissue section placement are calculated and compared. These estimates are used to train a statistical classifier to label each pixel within the capture area as either tissue or background.
The quality control image below shows the result of this process. Individual image pixels determined to be tissue are colored light blue. The locations of the invisible capture spots are drawn as gray circles when over background, or dark blue spots over tissue.
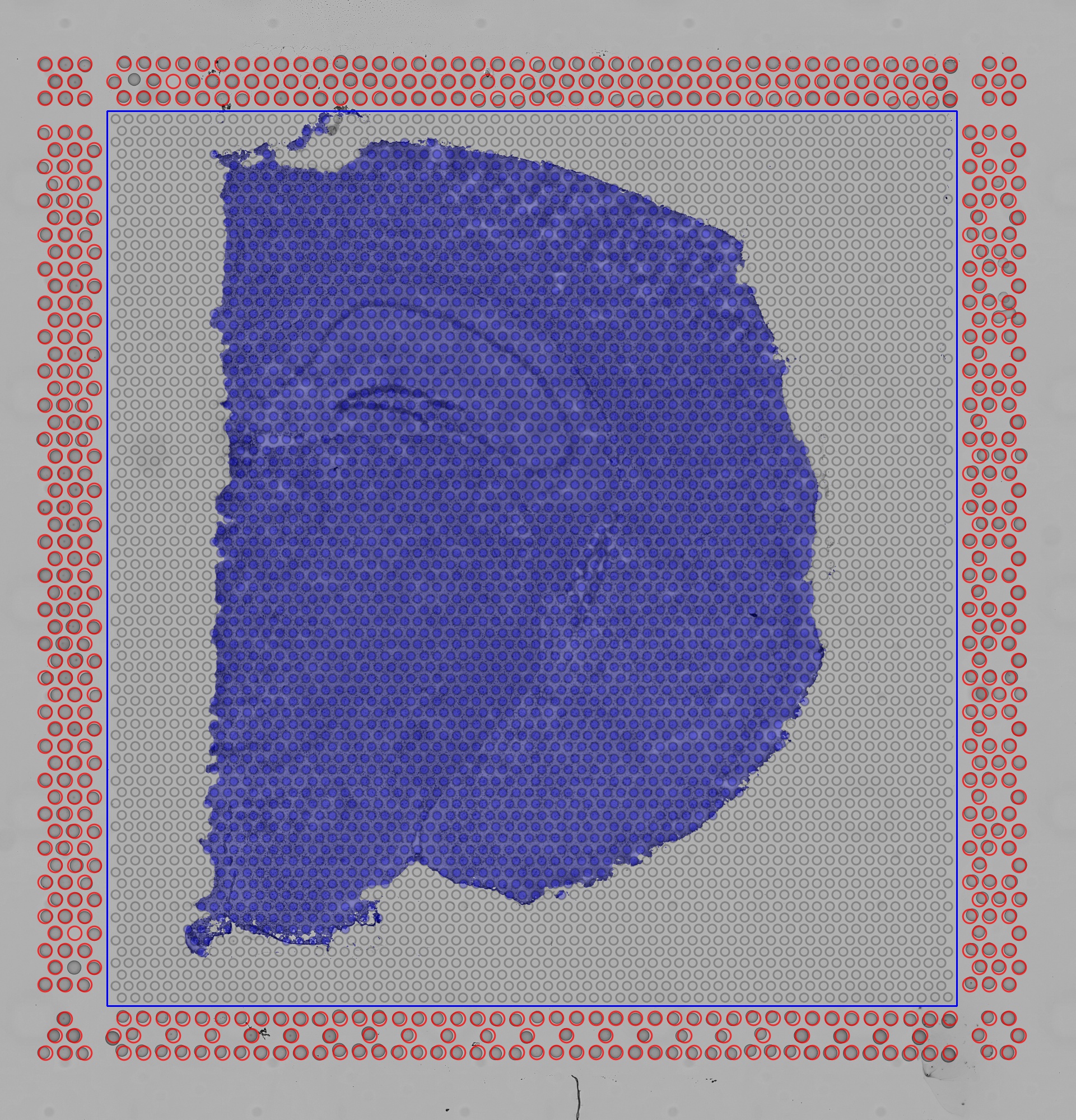 |
| Quality Control Image |