Cell Ranger6.0, printed on 04/02/2025
This page describes the output file structure from the cellranger multi subcommand specifically for 3' Cell Multiplexing data. This subcommand was introduced in Cell Ranger 5.0 for joint analysis of 5' gene expression and VDJ (GEX + VDJ) data, and in Cell Ranger 6.0 for 3' Cell Multiplexing data.
Upon completion, the cellranger multi subcommand will produce an outs directory with the following structure:
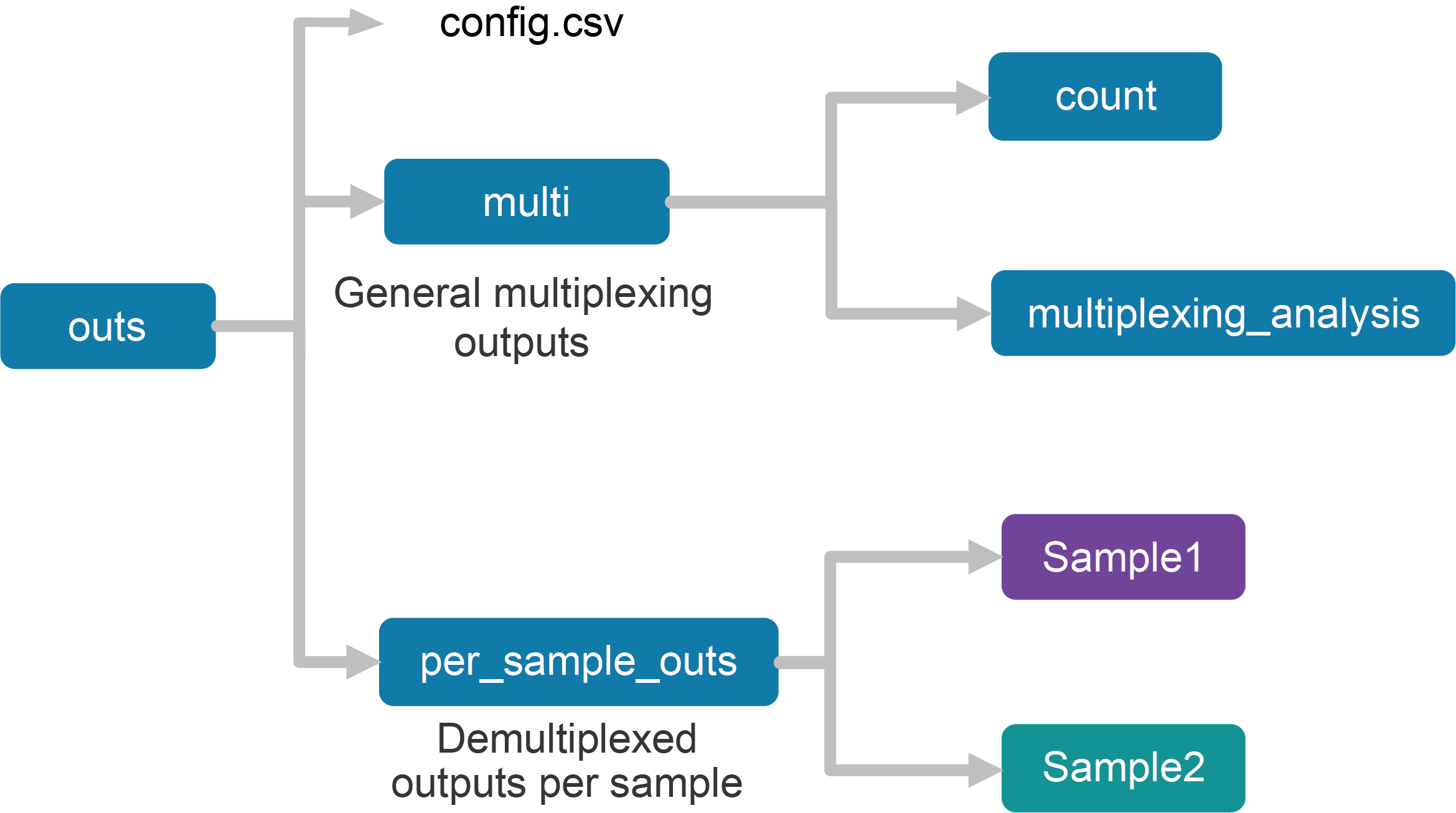
The files in the multi folder are generic to the entire multiplexing experiment, while the files in the per_sample_outs directory have been demultiplexed to single samples.
└─ multi
├── count
│ ├── feature_reference.csv
│ ├── raw_cloupe.cloupe
│ ├── raw_feature_bc_matrix
│ │ ├── barcodes.tsv.gz
│ │ ├── features.tsv.gz
│ │ └── matrix.mtx.gz
│ ├── raw_feature_bc_matrix.h5
│ ├── raw_molecule_info.h5
│ ├── unassigned_alignments.bam
│ └── unassigned_alignments.bam.bai
└── multiplexing_analysis
├── assignment_confidence_table.csv
├── cells_per_tag.json
├── tag_calls_per_cell.csv
└── tag_calls_summary.csv
The count folder contains files including:
The multiplexing_analysis folder contains files including:
These files are further described here.
The assignment_confidence_table.csv table provides a summary of all the information from the tag-assignment algorithm for each cell-associated barcode, including the probability that a given barcode belongs to a given state. The user may modify the confidence threshold used for assigning tags to barcodes on their own in a data-science environment like Python or R to enable further downstream analysis, if they so desire.
Row,CMO301,CMO302,Barcodes,Multiplet,Blanks,Assignment,Assignment_Probability 1,1.7492220542497986e-10,0.999994,AAACCCAAGATTCGAA-1,5.0927025002342424e-06,7.880223262070917e-07,CMO302,0.999994 2,0.9936742,1.95217130055954e-09,AAACCCAAGCAACAGC-1,0.0063257301365501695,7.021953108383088e-09,CMO301,0.9936742 13,2.3960595409731583e-08,0.0064328,AAACCCATCATTCGGA-1,0.993567110839031,3.554702514325053e-12,Multiplet,0.9935671 35,2.74875783854256e-12,0.00021014,AAACGCTGTCCAAGAG-1,1.3255406427923249e-17,0.9997898543154408,Blanks,0.999789 39,1.4235671575612072e-05,0.55488715,AAACGCTGTTCTTCAT-1,4.071660981663313e-07,0.44509820,Unassigned,0.55488715
Column descriptions:
The tag_calls_summary.csv summarizes multiplexing results by providing statistics about categories including the number of cells assigned no tag, one tag, more than one tag, etc. The category No tag assigned includes both cells that were considered Blanks and cells considered Unassigned.
Category,num_cells,pct_cells,median_umis,stddev_umis No tag molecules,0,0.0,None,None No tag assigned,386,2.9,None,None 1 tag assigned,12465,93.6,None,None More than 1 tag assigned,472,3.5,None,None CMO301,6437,48.3,3442.0,7988.3 CMO302,6028,45.2,3515.5,5167.2 CMO301|CMO302,472,3.5,12414.0,9696.0
Column descriptions:
The tag_calls_per_cell.csv file contains tag calls per cell, one line for each barcode.
cell_barcode,num_features,feature_call,num_umis AAACCCAAGCAACAGC-1,1,CMO301,16778 AAACCCAAGCTCGTGC-1,1,CMO301,1735 AAACCCACATGACTGT-1,1,CMO301,1625 AAACCCAGTCCACAGC-1,1,CMO301,19323 AAACCCAGTCGCGGTT-1,1,CMO301,1678
Column descriptions: