Cell Ranger3.1, printed on 04/01/2025
To reduce sequencing and library preparation costs, Feature Barcoding Only mode can be used. In Cell Ranger 3.1, cellranger countcan run using only Feature Barcoding libraries like an Antibody Capture library without sequencing the Gene Expression library.
To use cellranger in Feature Barcoding Only mode, follow instructions for Feature Barcoding Analysis, and omit Gene Expression entries from the Libraries CSV file. Results from Feature Barcoding Only analysis can be used with cellranger aggr, with the requirement that all aggregated runs must share a common feature reference.
Cell Ranger performs the normal Feature Barcoding quantification analysis in Feature Barcoding Only mode, but uses a simplified version of the cell calling algorithm. The EmptyDrops step that refines cell calls in not performed. See the Cell Calling algorithm documentation for details.
| We do not support analyzing libraries with peptide-MHC multimers only. We recommend using libraries with peptide-MHC mulitimers along with cell surface proteins targeting T-cell surface markers. |
| We also do not support analyzing a dataset with only a CRISPR Screening library. |
| If less than 10 antibodies are used in the Feature Barcoding only mode, Cell Ranger will skip secondary analysis and will not output a cloupe file. This restriction does not affect runs where Gene Expression libraries are present. |
A web_summary.html file summarizing the run is produced as usual. Metrics for Antibody libraries appear under the Antibody Sequencing and Antibody Application sections. The Sequencing and Mapping sections, which display metrics from Gene Expression libraries, do not appear.
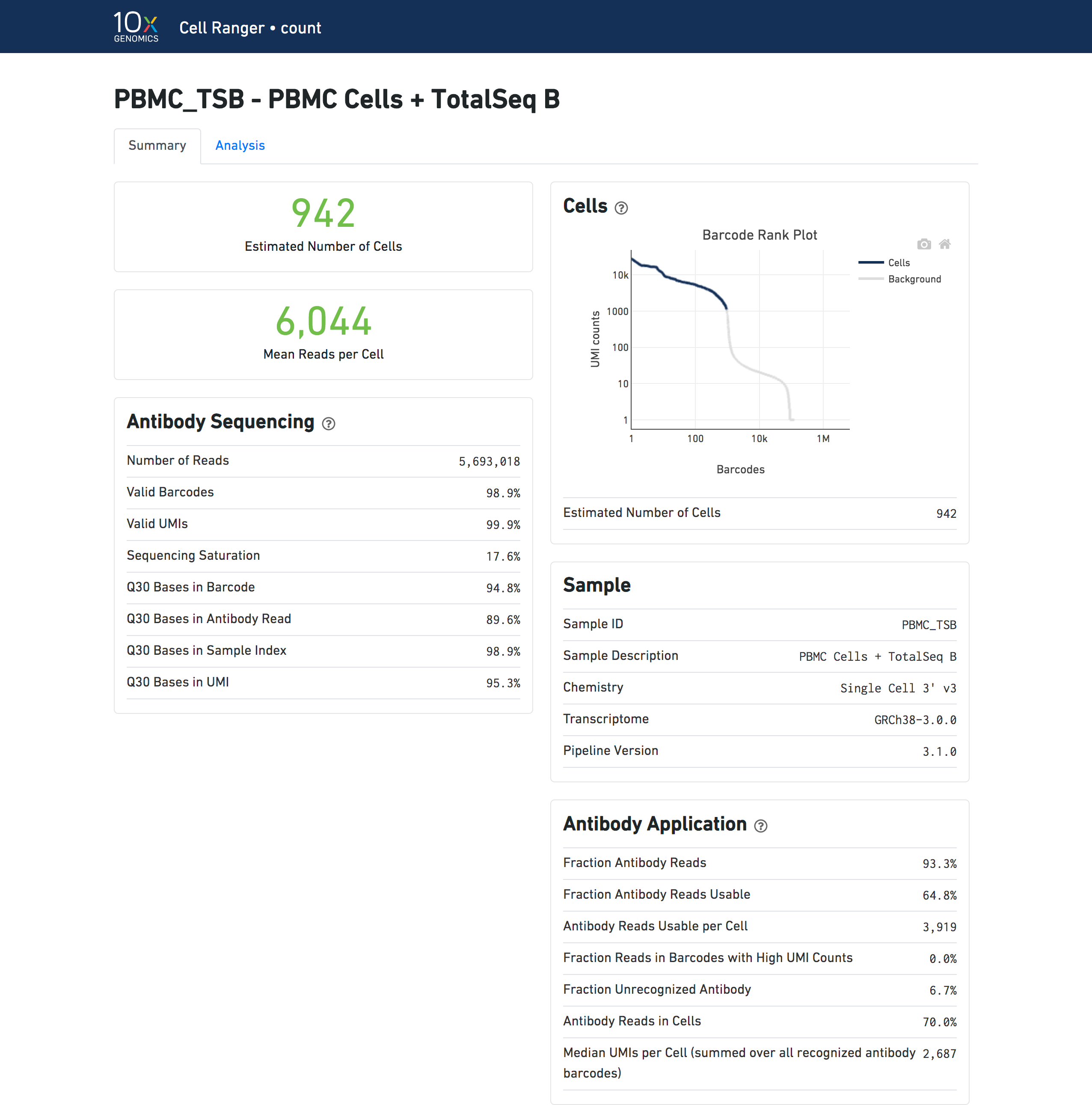
| Metrics results displayed in the web summary in Feature Barcoding Only mode do not precisely match results for Gene Expression + Feature Barcoding analysis, since there are differences in the set of cells that are called in each mode. |
Shown below are results for a single sample with Gene Expression and antibody Feature Barcoding libraries. On the left, both libraries are processed together, and the barcode-rank plot displays Gene Expression counts. On the right, only the Feature Barcoding library is supplied to cellranger count, and the barcode rank plot displays the antibody counts.
Observe that a similar number of cells were called in each cellranger run, but the shape of barcode-rank plot differs due to differences in the background levels of the Gene Expression and Antibody libraries.
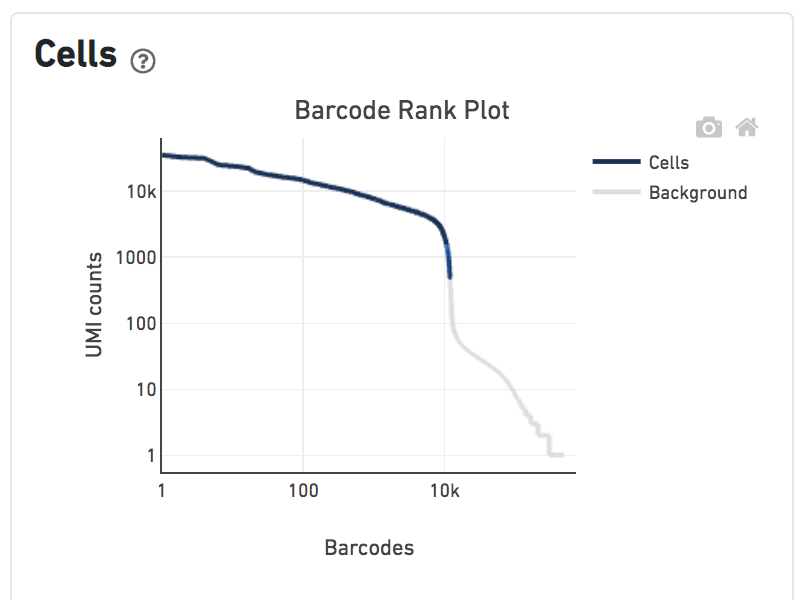
Gene Expression + Feature Barcoding |
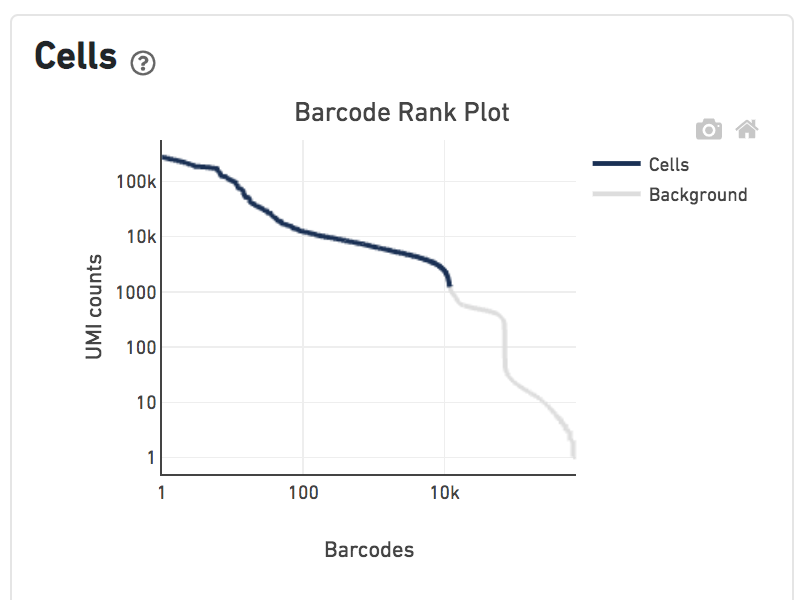
Feature Barcoding Only |
| The choice of antibodies in the panel plays a significant role in the effectiveness of cell calling in Feature Barcoding only mode. |
Feature Barcoding Only analysis only detects cells that are labelled by the Feature Barcoding reagents in use. Cell types not well-stained by the panel have low total UMI counts and may not be called by the cell-calling algorithm. The table below illustrates this effect by comparing the number of cell detected in the gene expression library and antibody capture library derived from the same cells. As the number of antibodies in the panel increases, the number of cells detected in the Feature Barcoding Only analysis approaches the number detected in the gene expression analysis. Below are results for a series of TotalSeq-B antibody panels applied to human PBMCs.
| Description | Loaded Cells | GEX Called Cells | Ab Called Cells | Called In Both |
|---|---|---|---|---|
| TotalSeq-B 4 Antibodies | 1000 | 705 | 562 | 533 (75.6%) |
| TotalSeq-B 5 Antibodies | 1000 | 749 | 653 | 626 (83.6%) |
| TotalSeq-B 14 Antibodies | 5000 | 4233 | 4389 | 3977 (93.9%) |
| TotalSeq-B 31 Antibodies | 5000 | 4616 | 4488 | 4540 (97.2%) |