Cell Ranger ATAC6.3, printed on 04/01/2025
Loupe Browser is a desktop application for Windows and MacOS that allows you to quickly and easily visualize and analyze Chromium™ Next Gem Single Cell ATAC data. It is optimized for finding significant peaks and distinguishing transcription factor motifs, identifying cell types, comparing chromatin accessibility between groups of cells, and exploring substructure within cell clusters.
Loupe is named for a jeweler's loupe, which is used to inspect gems.
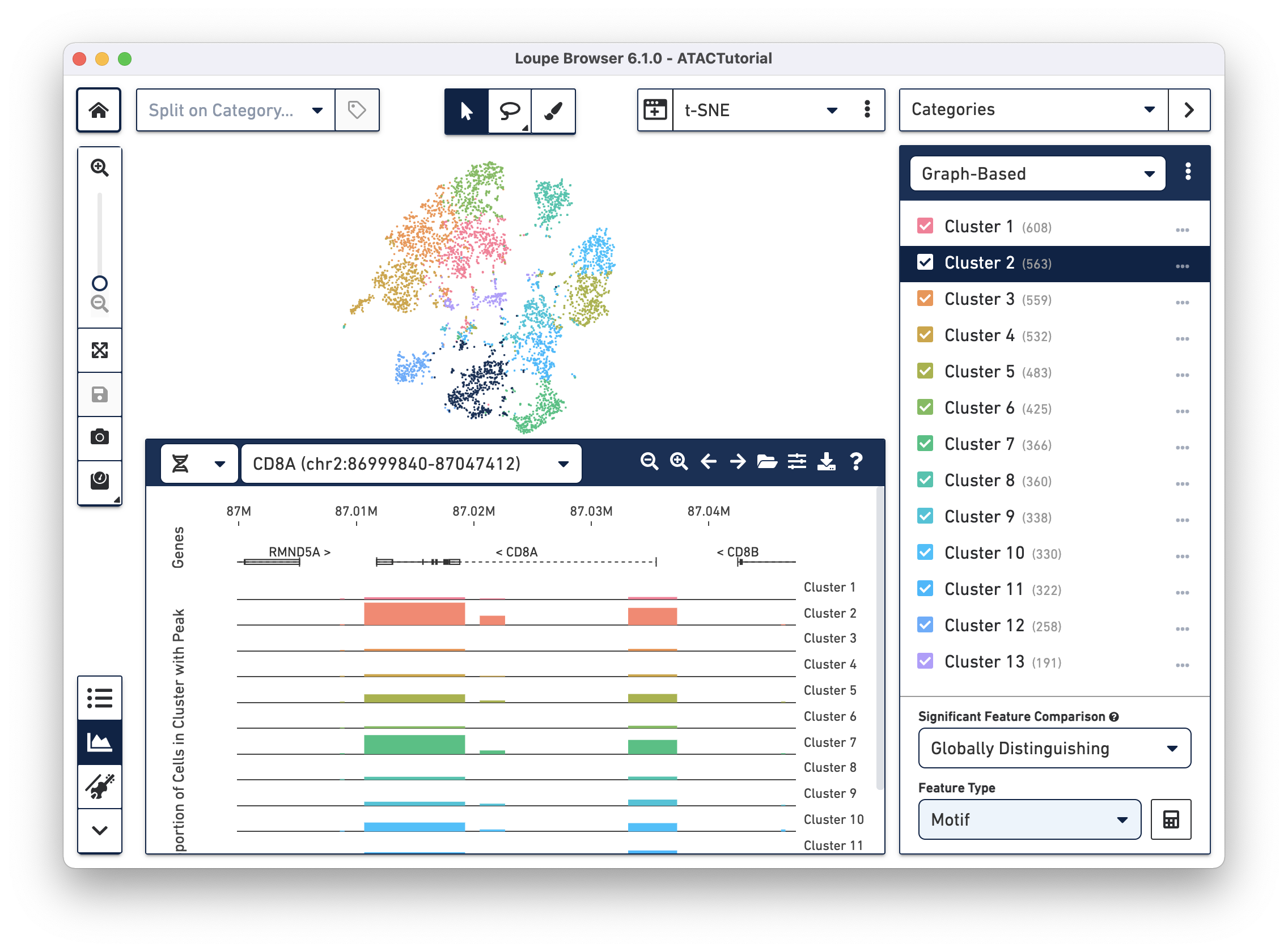
Loupe Browser opens .cloupe files generated by the Cell Ranger ATAC pipeline. Once your data is in Loupe Browser, you can rapidly explore and gain insights from the data without writing a single line of code.
Loupe Browser is built to accelerate the following applications:
Identifying Cell Types Find promoters and transcription factor motifs that differentiate distinct cell types and functional groups using Accessibility mode.
Exploring Cell Subtypes Use the filter panel to create complex boolean filters to find cell subtypes in your dataset.
Analyzing Differential Accessibility Use the ATAC Peak Viewer to pinpoint the location of differentially accessible genomic regions with putative regulatory function.
Finding Significant Features Create custom clusters and use differential accessibility tools to identify precise cell groups within your data.
Sharing Results Save features of interest, export data tables, and capture screenshots of your Single Cell ATAC data.
To walk through the features and uses of Loupe Browser for single-cell ATAC data, start the ATAC tutorial.
Loupe Browser can also be used to analyze Chromium™ Single Cell 5′ and 3′ Gene Expression, V(D)J, and Visium data. To learn how, consult the Loupe Browser for Gene Expression page.
Finally, if you have already used Loupe Browser to investigate Gene Expression data, and are new to ATAC, read Differences from Gene Expression.