Space Ranger1.3, printed on 03/10/2025
The pipeline output contains a subdirectory named spatial which stores imaging related files. These files are useful to confirm fiducdial alignment and tissue detection. However, the user supplied original full resolution images are used for visualization in Loupe Browser. The imaging files include the following:
tissue_hires_image.png and tissue_lowres_image.png: These files are downsampled versions of the original,
full-resolution brightfield image provided by the user. Downsampling is accomplished by box filtering, which averages
RGB values in patches of pixels in the full-resolution image to obtain an RGB value of one pixel in the downsampled
image. The tissue_hires_image.png image has 2,000 pixels in its largest dimension, and the tissue_lowres_image.png
has 600 pixels.
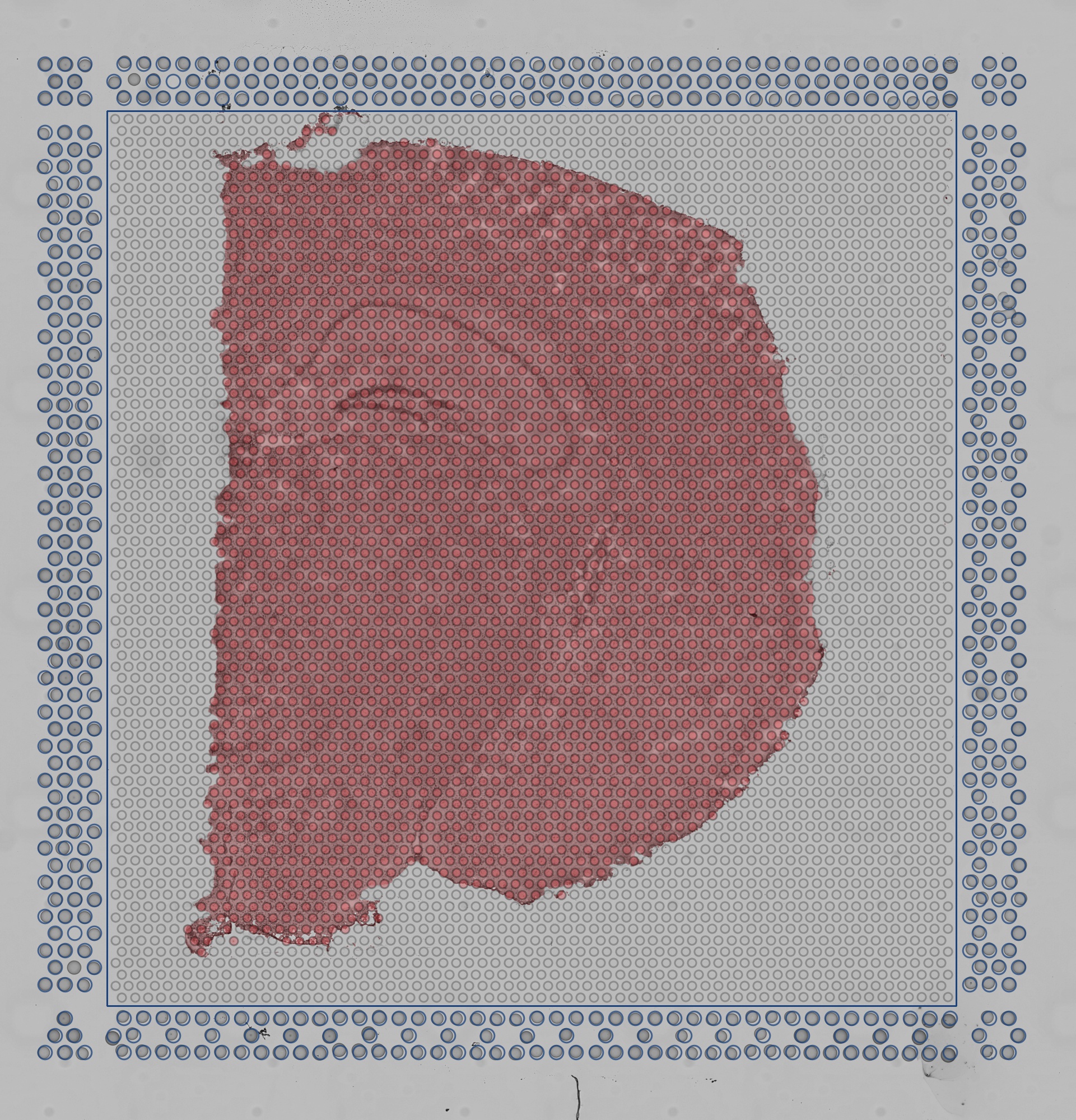
aligned_fiducials.jpg: This image has the dimensions of tissue_hires_image.png. Fiducial spots found by the
fiducial alignment algorithm are highlighted in red. This file is useful to verify that fiducial alignment was
successful.

scalefactors_json.json: This file contains the following fields:
tissue_hires_scalef: A scaling factor that converts pixel positions in the original, full-resolution image to
pixel positions in tissue_hires_image.png.tissue_lowres_scalef: A scaling factor that converts pixel positions in the original, full-resolution image to
pixel positions in tissue_lowres_image.png.fiducial_diameter_fullres: The number of pixels that span the diameter of a fiducial spot in the original,
full-resolution image.spot_diameter_fullres: The number of pixels that span the diameter of a theoretical 65µm spot in the original, full-resolution image.$ cd /home/jdoe/runs/sample345/outs/spatial $ cat scalefactors_json.json {"spot_diameter_fullres": 89.43834961021503, "tissue_hires_scalef": 0.17011142, "fiducial_diameter_fullres": 144.4773339857, "tissue_lowres_scalef": 0.051033426}
The example scalefactors_json.json values are derived from the adult mouse brain dataset which includes an image with dimensions of 11291 x 11757. The scalefactor is calculated as
$$scalefactor = \frac{\text{target size}}{max(\text{original image width, original image height})}$$
Since the largest dimension for the tissue_hires_image.png is 2000 pixels, the scalefactor will be $\text{tissue_hires_scalef} =\frac{2000}{11757} \approx 0.17011142$
Similarly for the tissue_lowres_image.png, the largest dimension is 600 pixels and the scalefactor is $\text{tissue_lowres_scalef} =\frac{600}{11757} \approx 0.051033426$
The spot diameter and fiducial diameter are estimated fractional pixels of the spot diameter in the original image. It is estimated based on the registration solution and the known size of the spots and fiducial frame and does not use a priori knowledge of the image pixel sizes. For this example the estimated pixel size was can be calculated from the spot diameter for the full resolution image $\text{microns/pixels} =\frac{65}{89.43834961} \approx 0.7267575965$
detected_tissue_image.jpg: This image has the dimensions of the tissue_hires_image.png and shows the following:
tissue_positions_list.csv : This text file contains a table with rows that correspond to spots. It has 4,992 rows, which is the number of spots in the spatial array. Columns, whose names are not specified in the file, correspond to the following fields:
barcode: The sequence of the barcode associated to the spot.in_tissue: Binary, indicating if the spot falls inside (1) or outside (0) of tissue.array_row: The row coordinate of the spot in the array from 0 to 77. The array has 78 rows.array_col: The column coordinate of the spot in the array. In order to express the orange crate arrangement of
the spots, this column index uses even numbers from 0 to 126 for even rows, and odd numbers from 1 to 127 for odd
rows. Notice then that each row (even or odd) has 64 spots.pxl_row_in_fullres: The row pixel coordinate of the center of the spot in the full resolution image.pxl_col_in_fullres: The column pixel coordinate of the center of the spot in the full resolution image.$ cd /home/jdoe/runs/sample345/outs/spatial $ column -s, -t < tissue_positions_list.csv | head -n 10 ACGCCTGACACGCGCT-1 0 0 0 1251 1210 TACCGATCCAACACTT-1 0 1 1 1371 1279 ATTAAAGCGGACGAGC-1 0 0 2 1251 1348 GATAAGGGACGATTAG-1 0 1 3 1371 1417 GTGCAAATCACCAATA-1 0 0 4 1251 1486 TGTTGGCTGGCGGAAG-1 0 1 5 1371 1554 GCATCCTCTCCTATTA-1 0 0 6 1251 1623 GCGAGGGACTGCTAGA-1 0 1 7 1371 1692 TGGTACCGGCACAGCC-1 0 0 8 1251 1761 GCGCGTTTAAATCGTA-1 0 1 9 1371 1830
The spots are arranged using a orange crate packing to maximize packing density. Further explanation of this arrangement can be found in this article.
Note that the tissue_positions_list.csv contains the same information about the barcode whitelist and their slide coordinates as that contained in the barcodes folder that comes bundled with Space Ranger (spaceranger-x.y.z/lib/python/cellranger/barcodes where x.y.z represent the version number).