Cell Ranger ARC1.0, printed on 04/07/2025
The pipeline output directory, described in Understanding Output, contains all of the data produced by one invocation of a pipeline (a pipestance) as well as rich metadata describing the characteristics of each stage. This directory contains a specific structure that is used by the Martian pipeline framework to track the state of the pipeline as execution proceeds.
Cell Ranger's notion of a pipeline is very flexible in that a pipeline can be composed of stages that run stage code or sub-pipelines that may themselves contain stages or sub-pipelines.
Cell Ranger pipelines follow the convention that stages are named with verbs (e.g., ALIGN_READS, MARK_DUPLICATES, FILTER_BARCODES) and sub-pipelines are named with nouns and prefixed with an underscore (e.g., _BCSORTER). Each stage runs in its own directory bearing its name, and each stage's directory is contained within its parent pipeline's directory.
For example, the cellranger-arc mkfastq pipeline has the following process graph:
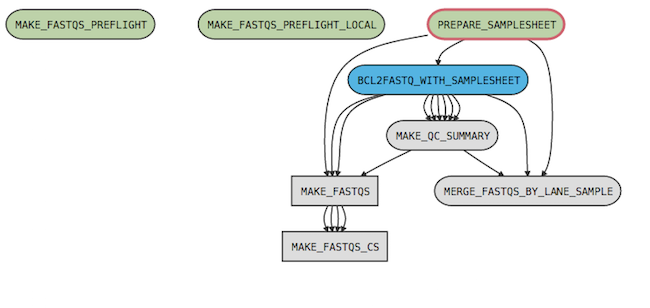
where
| The MAKE_FASTQS_CS stage is not strictly necessary since it contains no stages and only one child pipeline (MAKE_FASTQS); however, it serves to mask some of the low-level inputs required by the MAKE_FASTQS pipeline. |
Every pipestance operates wholly inside of its pipeline output directory. When the pipestance completes, this pipestance output directory contains three outputs: metadata files, the pipestance output file directory, and the top-level pipeline stage directory.
The top-level pipeline stage directory is a stage directory that contains any number of child stage directories as well as one stage output directory for each fork run by that stage. The top-level pipeline stages for Cell Ranger ARC are:
Most of the Cell Ranger ARC pipelines contain single-fork stages, which means there is one fork0 stage output directory within each stage directory. Chunk output directories are a subset of stage output directories that additionally contain runtime information specific to the job or process being run by that chunk (e.g., a process ID or cluster job ID).
For example, the cellranger-arc mkfastq pipeline's pipeline output directory contains the following directory structure:
| _log | Metadata file |
| outs/ | Pipestance output file directory |
| MAKE_FASTQS_CS/ | Top-level pipeline stage directory |
| MAKE_FASTQS_CS/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/fork0/files/ | Stage output files |
| MAKE_FASTQS_CS/MAKE_FASTQS/ | Stage directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/fork0/files/ | Stage output files |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/ | Stage directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/chnk0/ | Chunk output directory |
The metadata contained in the pipeline output directory includes
| File Name | Description |
|---|---|
| _finalstate | Metadata cache that is populated when a pipestance completes to minimize re-aggregation of metadata |
| _invocation | The MRO call used to invoke this pipestance |
| _log | The log messages that are reported to your terminal window when running cellranger-arc commands |
| _mrosource | The entire MRO describing the pipeline with all @include statements dereferenced |
| _perf | Detailed runtime performance data for every stage in the pipestance |
| _timestamp | The start and finish time for this pipestance |
| _vdrkill | A list of all of the volatile data (temporary files) removed during pipeline execution as well as total number of files and bytes deleted |
| _versions | Versions of the components used by the pipeline |
Stage directories contain stage output directories, stage output files, and the stage directories of any child stages or pipelines.
Stage output directories typically contain:
| File Name | Contents |
|---|---|
| files/ | Directory containing any files created by this stage that were not considered volatile (temporary) |
| split/ | A special stage output directory for the step that divided this stage's input into parallel chunks |
| chnkN/ | A chunk output directory for the Nth parallel chunk executed |
| join/ | A special stage output directory for the step that recombined this stage's parallel output chunks into a single output dataset again |
| _complete | A file that, when present, signifies that this stage has successfully completed |
| _errors | A file that, when present, signifies that this stage failed. Contains the errors that resulted in stage failure. |
| _invocation | The MRO call used to execute this stage by the Martian framework |
| _outs | The output files generated by this stage |
| _vdrkill | A list of all of the volatile data (temporary files) removed during pipeline execution as well as total number of files and bytes deleted |
Chunk output directories are a subset of stage output directories that, in addition to the aforementioned stage output, may contain:
| File Name | Contents |
|---|---|
| _args | The arguments passed to the stage's stage code |
| _jobinfo | Metadata describing the stage's execution, including performance metrics, job manager jobid and jobname, and process ID |
| _jobscript | The script submitted to the cluster job manager (cluster mode-only) |
| _stdout | Any stage code output that was printed to the stdout stream |
| _stderr | Any stage code output that was printed to the stderr stream |
These metadata files should be treated as read-only, and altering the contents of metadata files is not recommended.
Pipestance output directories can demonstrate very complicated structures, and re-attaching the Cell Ranger ARC UI is the easiest way to quickly navigate to a pipeline stage of interest and examine its metadata. In the absence of being able to access the UI, the standard find command can quickly return high-level information about a pipestance.
For example, to find the stages that resulted in the overall failure of a pipestance whose output directory is sample345/,
$ find sample345/ -name _errors sample345/SC_ATAC_GEX_COUNTER_CS/SC_ATAC_GEX_COUNTER/_ATAC_MATRIX_COMPUTER/_PEAK_CALLER/COUNT_CUT_SITES/fork0/chnk0-u308e57a713/_errors
This tells us that the failed stage was COUNT_CUT_SITES.
It can be helpful to view all _errors files' contents at once by piping to xargs cat:
$ find sample345/ -name _errors | xargs cat Traceback (most recent call last): File "/home/jdoe/cellranger-arc-1.0.0/external/martian/adapters/python/martian_shell.py", line 659, in _main stage.main() File "/home/jdoe/cellranger-arc-1.0.0/external/martian/adapters/python/martian_shell.py", line 618, in main self._run(lambda: self._module.main(args, outs)) File "/home/jdoe/cellranger-arc-1.0.0/external/martian/adapters/python/martian_shell.py", line 589, in _run cmd() File "/home/jdoe/cellranger-arc-1.0.0/external/martian/adapters/python/martian_shell.py", line 618, inself._run(lambda: self._module.main(args, outs)) File "/home/jdoe/cellranger-arc-1.0.0/mro/atac/stages/processing/count_cut_sites/__init__.py", line 119, in main contig=args.contig, filename=args.fragments, index=args.fragments_index File "/home/jdoe/cellranger-arc-1.0.0/lib/python/atac/tools/io.py", line 136, in parsed_fragments_from_contig fragments = pysam.TabixFile(filename, index=index) File "pysam/libctabix.pyx", line 351, in pysam.libctabix.TabixFile.__cinit__ File "pysam/libctabix.pyx", line 404, in pysam.libctabix.TabixFile._open File "pysam/libchtslib.pyx", line 516, in pysam.libchtslib.HTSFile.tell NotImplementedError: seek not implemented in files compressed by method 1
In the above case, the error is an unhandled exception that results in a stack trace and whose cause is not obvious; these sorts of failures should be reported to the 10x software support team for assistance with diagnosis. (Note: this failure was induced by truncating the fragments file while the pipestance was running.)
Stages whose stage code run external binaries (for example, the ALIGN_AND_COUNT stage which runs STAR) often generate output to their stdout and stderr streams. These messages are captured in the _stdout and _stderr metadata files within the chunk output directories, and combining find and xargs cat to examine their contents can also assist with troubleshooting.