Cell Ranger DNA1.1, printed on 04/03/2025
| Analysis software for the 10x Genomics single cell DNA product is no longer supported. Raw data processing pipelines and visualization tools are available for download and can be used for analyzing legacy data from 10x Genomics kits in accordance with our end user licensing agreement without support. |
Now that you have had a thorough walkthrough of this visualization tool, there might be other analyses you would like to perform with this data that are not part of Loupe scDNA Browser yet. There are many ways to conveniently export screenshots or data in an interchangeable format for further downstream analysis.
In the toolbar, there is a button with the icon of an arrow pointing downward that allows exporting of data. This section will cover the three options in that dropdown.
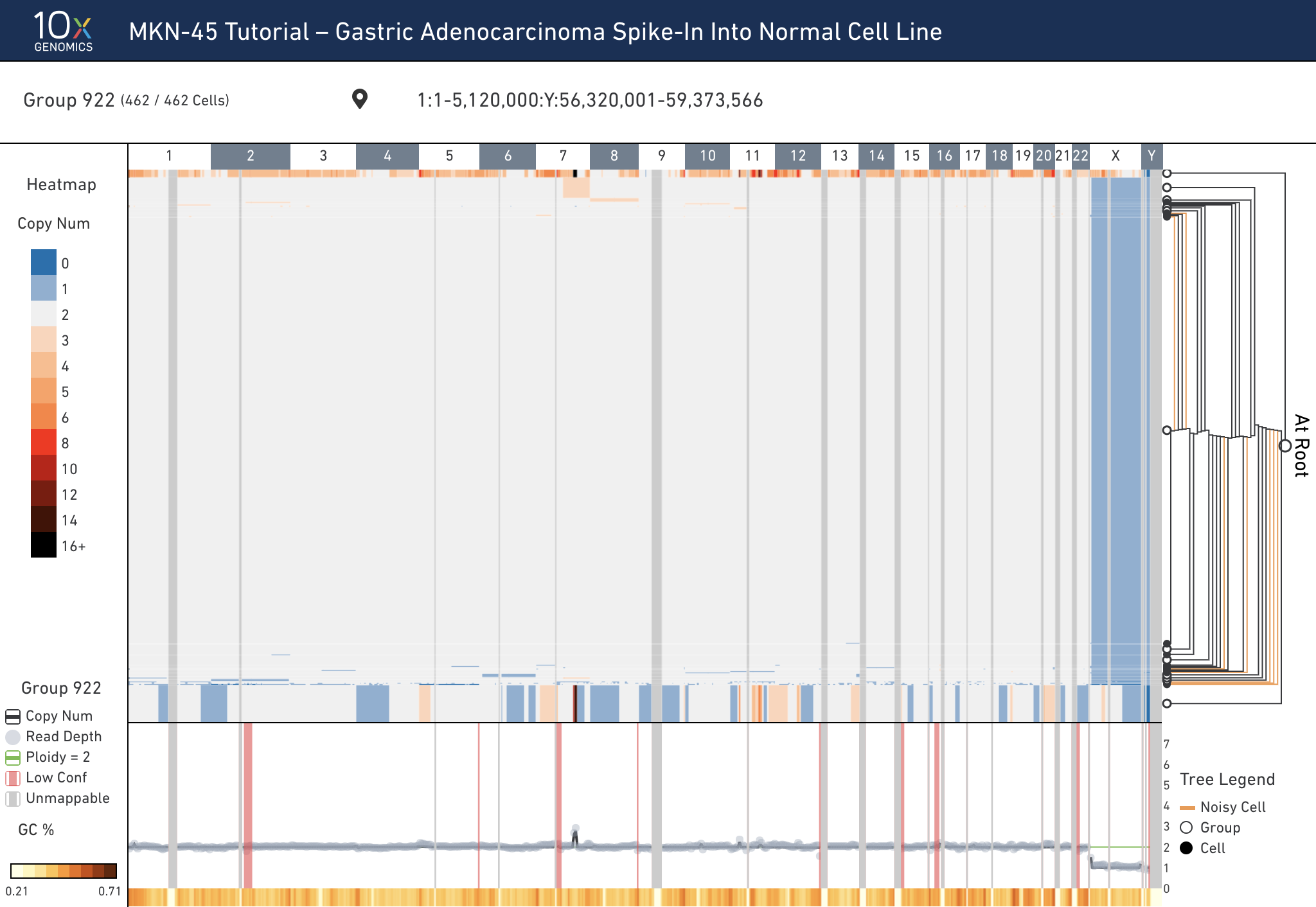
To export a screenshot of the interface as a PNG, select the first option in the export menu. This option will remove all of the interactive UI elements and produce a clean image of the data. Here is an example of what that looks like:
To export the currently displayed heatmap data as a CSV file, select the Heatmap Data as CSV. Depending on what's currently being displayed (either copy number or heterogeneity), the value for each bin will be exported. For each row in the heatmap, this option exports:
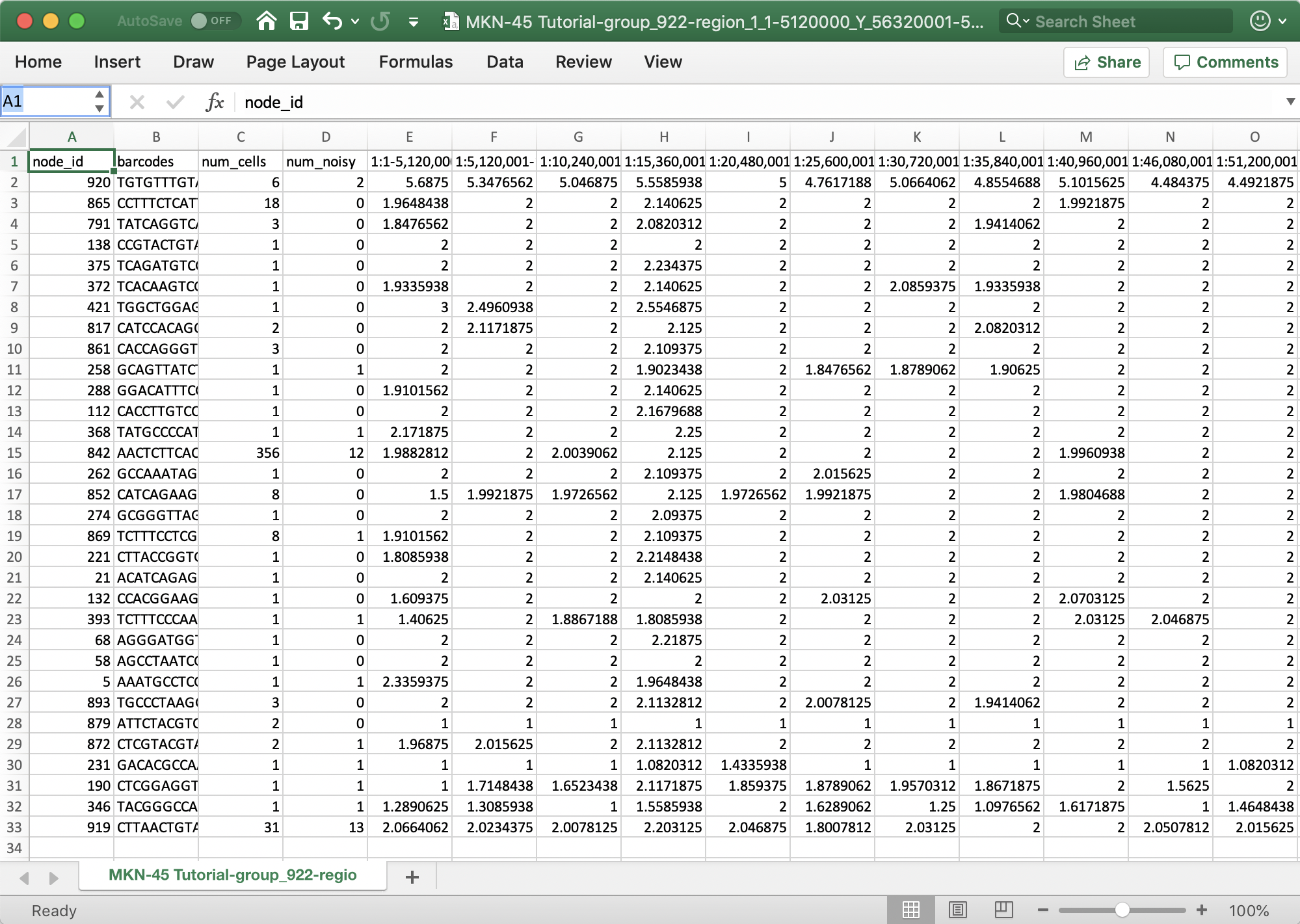
To export the data that results in the current trace, select the Group Data as CSV. In this exported CSV, the copy number, normalized read depth, heterogeneity, confidence, and GC content are exported for each bin. Remember, bins may be downsampled when zoomed out at larger genomic ranges.