Cell Ranger ARC2.0, printed on 04/11/2025
The cellranger-arc count pipeline outputs atac_possorted_bam.bam, a position-sorted and indexed BAM file for the Chromatin Accessibility library. This file is primarily provided for use with a BAM visualization tool such as the Integrated Genome Viewer (IGV).
| File | Records | Reference | Description |
| possorted_bam.bam | Reads | User-specified reference | Barcode-corrected reads aligned to the user-specified reference, sorted by reference position. |
The following assumes basic familiarity with the BAM format. More details on the SAM/BAM standard are available online.
Chromium cellular barcode and mapping information for each read is stored as TAG fields:
| Tag | Type | Description |
|---|---|---|
CB | Z | Chromium cellular barcode sequence that is error-corrected, confirmed against a list of known-good barcode sequences and translated. |
CR | Z | Chromium cellular barcode sequence as reported by the sequencer. |
CY | Z | Chromium cellular barcode read quality. Phred scores as reported by sequencer. |
BC | Z | Sample index read. |
QT | Z | Sample index read quality. Phred scores as reported by sequencer. |
TR | Z | Adapter sequence trimmed off the end of the read. |
TQ | Z | Base quality for the trimmed adapater sequence. Phred scores as reported by sequencer. |
The cell barcode CB tag includes a gem group suffix -1 that labels the GEMs
from a single well.
AGAATGGTCTGCAT-1
Cell Ranger ARC currently only supports libraries generated from a single GEM run
and so the gem group suffix is always -1. It can either be left in place and
treated as part of a unique barcode identifier, or explicitly parsed out to
leave only the barcode sequence itself.
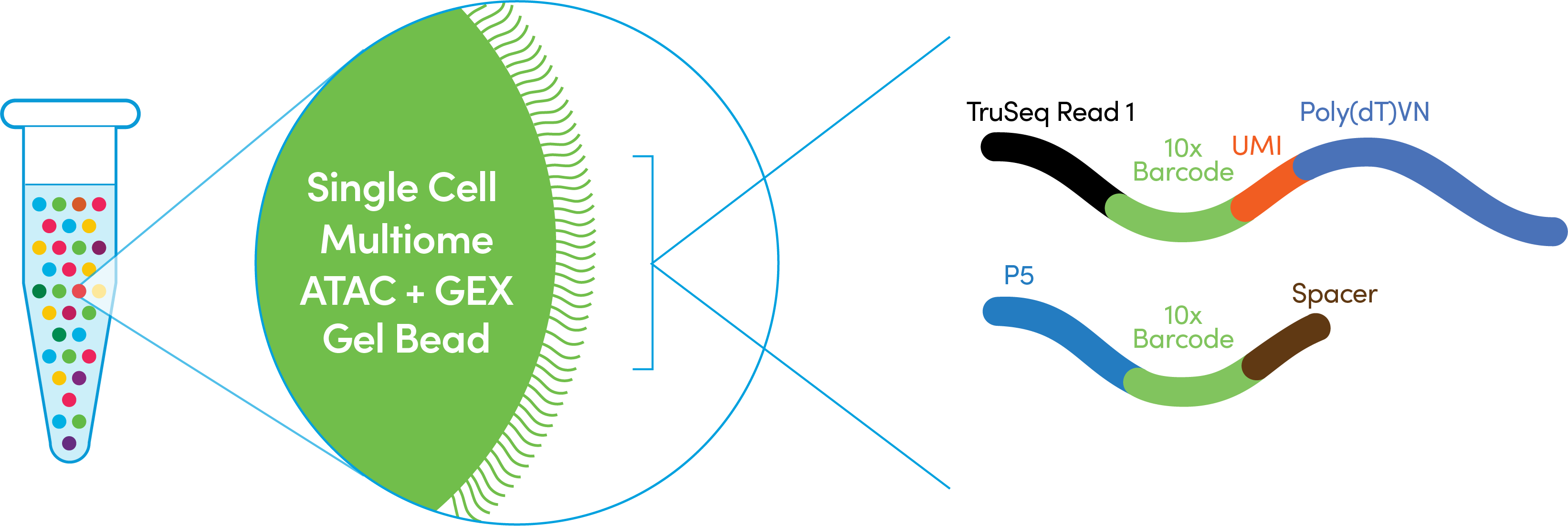
Chromium Single Cell Multiome ATAC + Gene Expression Gel Beads include a poly(dT) primer that enables production of barcoded, full-length cDNA for Gene Expression (GEX) library and a Spacer primer that enables barcode attachment to transposed DNA fragments for Chromatin Accessibility (ATAC) library. The 10x Barcode sequences on the ATAC and GEX specific primers on the same Gel Bead are not identical. Each Gel Bead has a unique pairing of ATAC and GEX specific barcode sequences. Barcode translation refers to the in silico translation of the error-corrected ATAC barcode as reported by the sequencer to its corresponding paired GEX barcode sequence.